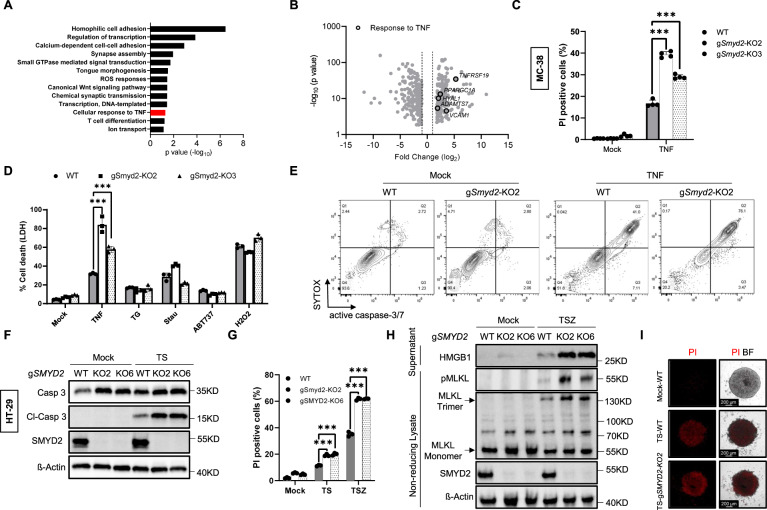
Fig. 3. SMYD2 deficiency sensitizes CRC cells to TNF-induced apoptosis and necroptosis.
A Gene ontology analysis for biological processes among the significantly and commonly regulated genes between the two clones of SMYD2 deficient HT-29 implanted tumors compared against WT HT-29 implanted tumors. Cellular response to TNF (highlighted red) among the significantly upregulated biological processes. B Volcano plot showing the expression of significantly downregulated and upregulated genes in SMYD2 deficient tumors compared to WT tumors. Black circles highlight genes related to TNF response. C Flow cytometric quantification of Propidium iodide (PI) stained WT and two different clones of Smyd2 deficient MC-38 cells stimulated with 0.1 ng/ml TNF or vehicle (mock). D LDH release from WT and Smyd2 CRISPR/Cas9 knockout MC-38 cells stimulated with vehicle (Mock), 0.1 ng/ml TNF, 25 µM thapsigargin (TG), 5 µM staurosporine (Stau), 50 µM ABT737, or 0.5 mM H2O2 for 24 h. E Flow cytometric detection of cleaved caspase-3/7 activity and SYTOX from the indicated genotypes of MC-38 cells stimulated with 0.1 ng/ml TNF. F Immunoblot analysis of SMYD2, caspase 3, and Cl-caspase 3 in HT-29 cells treated with or without 40 ng/ml TNF + 4 nM LCL161 (TS). β-Actin served as a loading control. G Flow cytometric quantification of PI stained HT-29 cells stimulated with TS or 10 ng/ml TNF + 1 nM LCL161 + 20 µM Z-VAD (TSZ). H Immunoblot analysis of HMGB1 from the supernatants and pMLKL/MLKL from non-reducing cell lysates. WT and SMYD2 CRISPR/Cas9 knockout HT-29 cells were stimulated with or without TSZ. β-Actin served as a loading control. I Representative images of PI stained HT-29 cells that were cultured in 3D. WT and SMYD2 CRISPR/Cas9 knockout HT-29 cells were stimulated with or without TS. DMSO was used as control. Experiments were performed three times and representative data are shown. Data are presented as mean +SD and student’s t-test was used for statistical calculation. **P < 0.01 and ***P < 0.001. N.S. not significant.