Fig. 1.
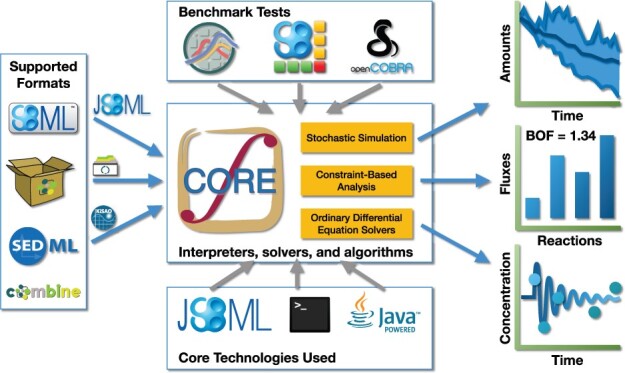
The capabilities of the SBSCL as an overview. Supported input model definitions include SBML, possibly with an experiment configuration file (SED-ML) or bundled in a package file (OMEX). The model is parsed using the JSBML library, and solutions are numerically computed for the corresponding ODE or SDE system over time, following the specified constraints and algorithm (e.g., Rosenbrock, Euler, Gillespie) or via linear programming. Once the simulation completes, the model results are reported either graphically using a line plot or tabular form. The results can be exported to formats such as CSV for downstream use. For testing the library, its implementation, its robustness, reliability, and efficient reproducibility of the results, open model collections such as BiGG (Norsigian et al., 2019) and BioModels (Malik-Sheriff et al., 2020) are utilized, which comprise several hundred SBML models and their SED-ML configurations
