Abstract
Objective:
To determine if a handheld, nanopore-based DNA sequencer can be used for rapid preimplantation genetic screening (PGS).
Design:
Laboratory study.
Setting:
Academic medical center.
Patient(s):
Amplified genomic DNA from euploid and aneuploid trophectoderm biopsy samples (n=9) that was also tested using traditional next generation sequencing (NGS).
Intervention(s):
Short-read DNA library preparation and nanopore-based sequencing using a hand-held MinION sequencer.
Main Outcome Measure(s):
Comparison of cytogenetic testing result from NGS and nanopore-based sequencing and the time required for library preparation and sequencing.
Result(s):
Multiplexed short-read DNA library preparation was completed in 45 minutes. Sequencing on a single sample was completed within 20 minutes and 5 samples were simultaneously sequenced in under 2 hours. Whole-chromosome aneuploidy screening results obtained from nanopore-based sequencing were identical to those obtained using NGS.
Conclusion(s):
Here we report the first application of nanopore-based sequencing for PGS on trophectoderm biopsy samples using a novel rapid multiplxed short-read nanopore sequencing library preparation protocol. Sequencing for aneuploidy screening could be performed on a single sample in 20 minutes and on 5 samples, simultaneously, within 2 hours. Overall, nanopore sequencing is a promising tool to perform rapid PGS onsite, enabling same day testing and embryo transfer, thus obviating the need for complex, large and expensive DNA sequencers or embryo freezing.
Keywords: Rapid preimplantation genetic screening, handheld nanopore sequencer, onsite assay, aneuploidy screening
Abstract
Objetivo:
Determinar si la secuenciación manual de ADN basado en nanoporos puede ser utilizada para el cribado genético preimplantacional rápido.
Diseño:
ESTUDIO retrospectivo.
Lugar:
Centro médico académico.
Paciente (s):
Muestras de ADN ampliado procedentes de biopsias de trofoectodermo tanto euploides como aneuploides (n=9), que fueron analizadas según la técnica de secuenciación masiva clásica (NGS).
Intervenciones:
Preparación de la librería de DNA y secuenciación manual de ADN minION basado en nanosporos.
Medidas prinicpales:
Comparación de los resultados citogenéticos obtenidos tras NGS y secuenciación basada en nanosporos y el tiempo empleado para la preparación de la librería y la secuenciación.
Resultados:
La preparación de la librería de ADN de lectura corta multiplexada se completó en 45 minutos. La secuenciación de una muestra aislada se complete en 20 minutos y 5 muestras pudieron ser secuenciadas simultáneamente por debajo de 2 horas. Los resultados del cribado de aneuploidias de todos los cromosomas obtenidos por la secuenciación manual de ADN basado en nanoporos fueron idénticos a aquellos obtenidos mediante NGS.
Conclusiones:
Aquí se informa de la primera aplicación de la secuenciación manual de ADN basada en nanoporos en muestras de biosia de trofoectodermo para cribado genético preimplantacional utilizando una librería de preparación rápida y multiplexada. La secuenciación para un cribado de aneuploidias de una muestra aislada pudo realizarse en 20 minutos y 5 muestras pudieron ser secuenciadas simultáneamente por debajo de 2 horas. En GENERAL, la secuenciación por nanoporos es una herramienta prometedora para realizar el análisis de PGS in situ de manera rápida y con un tiempo de respuesta rápido, permitiendo en el mismo día el test y la transferencia embrionaria y con ello, obviando la necesidad de utilizar secuenciadores largos y complejos o congelar embriones.
Human embryos have high rates of chromosomal aneuploidy, a significant cause of implantation failure and spontaneous pregnancy loss (1, 2). Preimplantation genetic screening (PGS) is an effective method to assess the numerical chromosomal constitution of embryos prior to transfer in patients undergoing in vitro fertilization (IVF). PGS facilitates the selection of euploid embryos for transfer, thereby improving implantation rates and reducing the rates of miscarriage (3–5). By improving implantation rates in IVF, PGS empowers the practice of elective single embryo transfer (6, 7) thereby avoiding the increased obstetrical complications resulting from multiple gestation which occurs frequently after multiple embryo transfer.
Initially, PGS involved fluorescent in-situ hybridization (FISH) to assess the ploidy status of a subset of chromosomes. Likely due to limitations of the technology and the impact of blastomere biopsy at the cleavage stage, FISH-based PGS was shown to not be beneficial (8). Subsequent technologies included array-based comparative genomic hybridization, single nucleotide polymorphism microarray and quantitative real time polymerase chain reaction that enabled analysis of all chromosomes. These methods were limited by their high cost per sample and low throughput (9). Over the past several years, second-generation sequencing technologies, also known as next-generation sequencing (NGS), consisting of massively-parallel, high-throughput methods utilizing sequencing- by-synthesis-based chemistries have become increasingly widely used due to their higher dynamic range, lower cost, and ability to detect mosaicism (10–15). However, NGS-based aneuploidy detection still requires the use of complex and costly DNA sequencers and requires >12 hours for library preparation and sequencing, thereby necessitating embryo cryopreservation and often requiring the use of specialized reference laboratories.
Recently, third-generation sequencing technologies based on single-molecule sequencing have been developed. A prominent example is nanopore-based sequencing in which protein nanopores, approximately 10 nm in diameter, are distributed across a flow-cell membrane (16). An electrical current drives single-stranded DNA through the nanopore and the voltage changes that occur as each nucleotide passes through the nanopore are recorded (16). The identity of the nucleic acid is determined based on the variable resistivity of each base. The first commercial sequencer using nanopore technology is the MinION, which was released by Oxford Nanopore Technologies in 2014. The MinION is an 87 gram hand-held device that is the size of a deck of cards and is powered by a USB cable attached to a computer. Compared to traditional NGS technologies, nanopore-based sequencing has the distinct advantage of sequencing 15,000 times faster, delivering sequencing results in real-time, and a 99× lower capital equipment cost (~$1,000 for a MinION startup package vs. >$99,000 for a MiSeq sequencer).
We developed a method for rapid library preparation and sequencing of short DNA fragments and demonstrated that this method can provide rapid, array-based comparative genomic hybridization-concordant aneuploidy detection from chorionic villus cells retrieved from products of miscarriage (17). We then developed a method to barcode and multiplex multiple samples within a single sequencing run (18). NGS technologies typically require days in order for results to become available, precluding a fresh embryo transfer when a biopsy is obtained at the blastocyst stage of development. The speed of the MinION based approach would provide PGS results within a few hours and therefore allow for potential fresh embryo transfer, avoiding the need for freeze-all cycles when PGS is employed.
In the present study, we apply our MinION rapid multiplexed short-read sequencing technology to aneuploidy detection in PGS and demonstrate that our optimized MinION library preparation and data analysis protocol is concordant to Illumina VeriSeq PGS aneuploidy results from DNA obtained after trophectoderm biopsy.
MATERIAL AND METHODS
Samples
Trophectoderm 3–5 cell biopsy samples obtained from fresh day 5 embryos as part of routine clinical care were sent to a reference lab (combimatrix) for routine clinical PGS testing. Samples were subjected to SurePlex whole genome amplification (WGA) and testing using the VeriSeq PGS assay following the manufacturer’s instructions as part of routine clinical care. This results in excess DNA that is not needed for routine clinical testing and that we used for our study. Excess DNA from each sample was de-identified and was then sent to our research laboratory for MinION library preparation and sequencing. Samples (n=9) were included in this laboratory study. The study was approved by the institutional review board at Albert Einstein College of Medicine and Columbia University Medical Center.
Library Preparation
For each sample, ~260 ng WGA amplified products were subjected to the MinION PGS assay using rapid multiplex library preparation according to our recently developed protocol (Fig. 1) (18). The full protocol is included in the Supplemental Methods. Blinded samples (n=9) consisting of diploid and aneuploid specimens, and one reference normal male sample, were tested using two MinION sequencing runs on the MinION MIN106 flowcell (Oxford Nanopore, MIN106). Each sequencing run consisted of one barcoded known reference sample and four blinded test samples. Initially a 2-hour sequencing run was performed using MINKNOW software version 1.7, and data were used for data analysis for aneuploidy detection. Additional 1-hour and 4-hour sequencing runs were performed after the initial sequencing run for mosaic aneuploidy and large copy number variation (CNV) detection.
FIGURE 1.

Library preparation for rapid nanopore sequencing. The workflow includes a standard ~2.5-hour SurePlex whole genome amplification, ~18 minute rapid end-preparation and purification, a 1-step barcode and sequencing adapter ligation and purification, and a 20-minute to 2-hour nanopore sequencing run, depending on the number of samples multiplexed into a single run. Components used in library preparation were specified in colors (gray, DNA fragments; red, native barcoding adapter; blue and orange, 1D sequencing adapter).
Data Analysis
Sequences were translated locally to fastq from fast5 format by MINKNOW local basecaller and Albacore. Fastq format sequences were converted to fasta format and subjected to cutadapt v1.14 (19) for de-multiplexing and short read removal using a ≥ 20 bp overlap to the adapter sequence, at an error rate of 0.10, and with a minimum read length of 50 bp after adapter trimming (parameter setting of −0 20 −e 0.10 −m 50). The de-multiplexed sequences from each sample were subjected to parallel blat (pBlat) alignment to human reference genome GRCh37 and uniquely mapped reads were screened by pslReps from the UCSC suite (20) using parameter −minCover=0.40, −minAli=0.80, −nearTop=0.001and –singleHit (17, 18). Numbers of sequences assigned to each chromosome were subjected to a modified Z-score method for aneuploidy detection as described in our previous studies (17, 18). Chromosomes with ≥ 25% copy number changes compared to a normal male reference were considered abnormal chromosomes, and they were eliminated in calculation of standard deviation for normal chromosomes to increase the detection sensitivity for WGA PGS samples. Statistical analysis was performed in R v3.4.0 (21). Samples had been de-identified, and the researcher was blinded to the VeriSeq PGS results at aneuploidy screening. A known normal male sample was included in each run as a reference.
For detection of CNV (≥ 20 Mb), 60,000 reads were used. Large CNVs were detected by segregating human reference genome into 10 Mbs bins, and aneuploidy detection was performed for each bin as described above. Bins with fewer than 100 uniquely assigned (UA) reads in the normal male reference samples were eliminated from large CNV detection assay. For bins with ≥ 200 UA reads in the reference sample, bins with Z-score > 3.29 were considered as a gain in CNV, and bins with Z-score < −3.29 was considered as a loss in CNV (P=.001); for bins with < 200 UA reads in the normal male reference sample, bins with Z-score > 6 were considered as gain in CNV, and bins with Z-score < 6 were considered as loss in CNV (P<.0001). Two bins with | Z-score | > 3.29 were concatenated. Large CNVs ≥ 20 Mbs were reported.
60K UA reads were used for an investigation of possibility of mosaic aneuploidy detection. The mosaic aneuploidy in a 5-cell biopsy can range from 20–80%, and a mosaic aneuploidy in a 3-cell biopsy can range from 33–67%. Chromosomes with ≥ 15% copy number changes (~30% mosaicism) compared to a normal male reference were arbitrarily considered as mosaic aneuploidy with high confidence and not included in calculation of the standard deviation of normal chromosomes to increase detection sensitivity for WGA PGS samples.
RESULTS
Sequencing Performance
Two nanopore sequencing runs, each comprised of five barcoded samples, were performed. Samples included 3 normal samples (two normal males and one normal female); 2 samples with aneuploidy of a single chromosome (one female monosomy 22, one female trisomy 19), and 4 samples with aneuploidy on more than one chromosome (a female with trisomy 22 and monosomy X, a female with trisomy 13 and monosomy 14, a male with XXY and trisomy 15, a female with mosaic trisomy 6, trisomy 15 and monosomy 18) (Table 1). Sample 6 consists of aneuploidy on chromosome (chr) 13 and chr15 and a large CNV gain on chr 15 (78914003–101997386) (Table 1). In Run1, 74,001; 144,129; 288,489; and 548,166 reads were generated in 15, 30, 60, and 120 minutes, respectively. In Run2, 89,027; 171,520; 347,397; and 664,769 reads were generated in 15, 30, 60, and 120 minutes, respectively (Fig. 2A, Table 1). The rate of sequencing was 274K-332K reads per hour through the testing. Of reads generated, >70% were able to be assigned to a unique barcode. Of the assigned reads, 75% to 95% reads from each sample were UA to the genome reference library (Hg19) (Table 1). Only those reads that were assigned to a barcode and uniquely matched to the reference genome library were used for subsequent downstream data analysis.
TABLE 1.
Performance of PGS using nanopore sequencing.
| Sample | Run | Barcode | Reads generated in first 2h | Reads generated in 3rd h | UAa | Full Chromosome CNVs | MinION assay result (using 30K UA reads) |
|---|---|---|---|---|---|---|---|
| 1 | 1 | NB02 | 56,991 | NA | 39,842 | Female, monosomy22 | Female, monosomy 22 |
| 2 | 1 | NB03 | 34,161 | NA | 31,799 | Female, trisomy 19 | Female, trisomy 19 |
| 3 | 1 | NB04 | 49,673 | NA | 40,180 | Female, trisomy 22; monosomy x | Female, trisomy 22; monosomy X |
| 4 | 1 | NB06 | 75,770 | NA | 40,121 | Normal male | Normal male |
| 5 | 1 | NB07 | 42,579 | NA | 40,087 | Normal female | Normal female |
| 6b | 2 | NB02 | 70,549 | 32,866 | 51,878 | Female, trisomy 13, monosomy 14 | Female, trisomy 13, monosomy 14 |
| 7 | 2 | NB03 | 44,343 | 19,904 | 41,742 | Female, mosaic trisomy 6; trisomy 15; monosomy 18 | Female, Trisomy 15; Monosomy 18c |
| 8 | 2 | NB04 | 58,247 | 25,583 | 52,040 | Normal male | Normal male |
| 9 | 2 | NB06 | 94,355 | 43,822 | 51,889 | Male, XXY sex chromosome complement, trisomy 15 | Male, XXY sex chromosome complement, trisomy 15 |
| 4 | 2 | NB07 | 54,840 | 24,421 | 51,910 | Normal male | Normal male |
Note: CNV = copy number variation; NA = not available; UA = uniquely assigned.
The first 42,500 reads from each samples in run1 and the first 55,000 reads in each sample in run2 were subjected to aligner to generate UA reads.
Sample 6 is a female, trisomy 13, monosomy 14, and large CNV gain on chromosome 15 (78,914,003–101,997,386) as detected by VeriSeq. The large CNV gain was not significant using 30K reads; it was significant using 60K UA reads.
The mosaic gain on chromosome 6 was not significant using 30K reads; it was significant using 60K UA reads.
FIGURE 2.
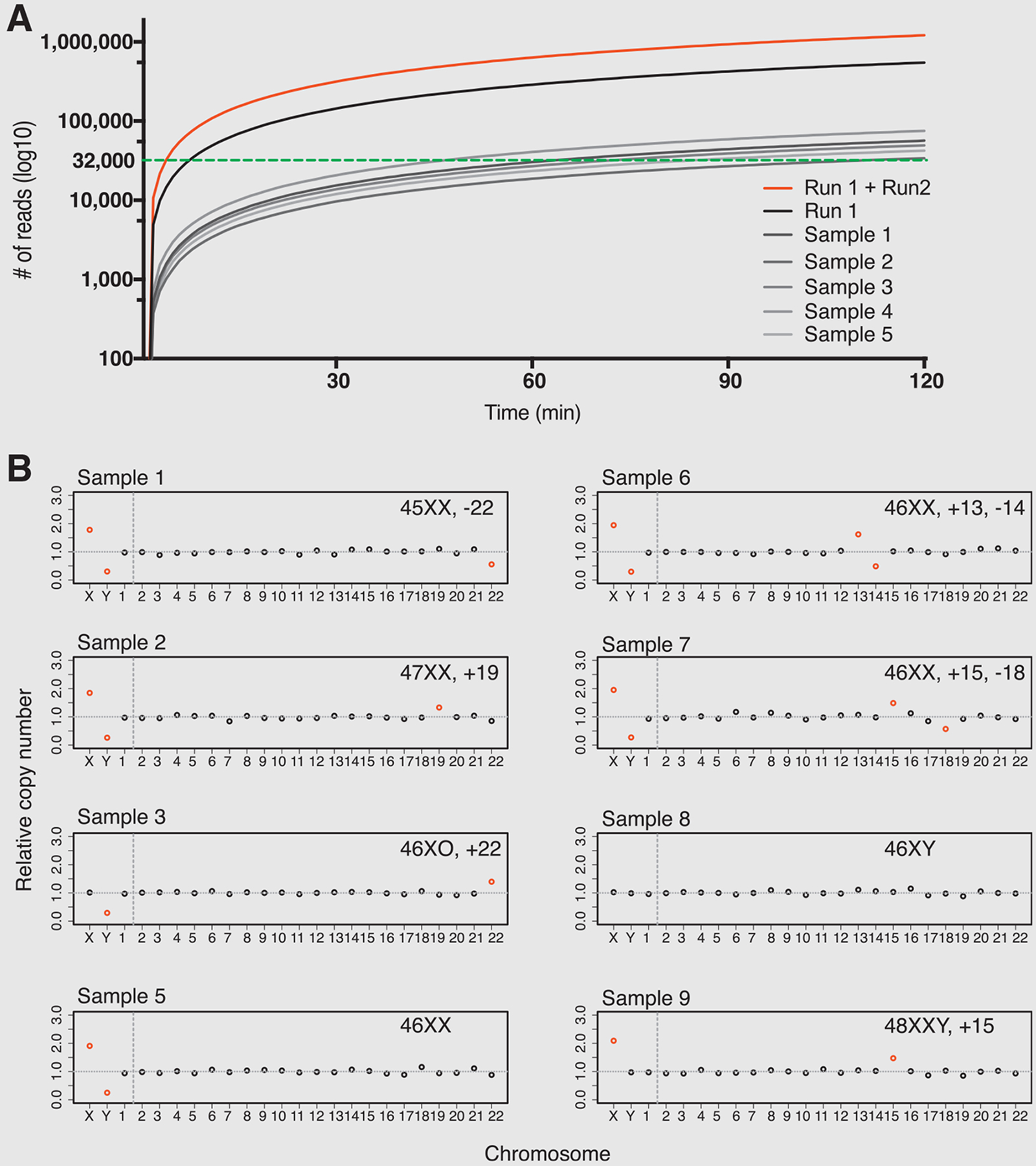
MinION assay results. (A) MinION nanopore sequencing representative yield performance (grayscale, sample 1–5; black, run 1 accumulative; red, run 1 and run 2 accumulative; green dotted line, threshold for ≥ 30K uniquely aligned reads for downstream analysis). (B) MinION-assisted aneuploidy detection results. The abnormality of each chromosome was indicated by color (red, significantly abnormal; black, normal).
Aneuploidy Detection
Aneuploidy detection analyses were performed based on Z-score method using the UA reads from each sample generated within 2-hour nanopore sequencing. Based on the known bias introduced by WGA, more reads are needed for aneuploidy detection assay when comparing to aneuploidy detection assay on samples that did not undergo WGA. Using the Z-score based assay with incremental read-intervals of 10,000 reads, we found that for whole aneuploidy detection a threshold of 30,000 reads was required. In Run 1, 48 to 112 minutes sequencing time was needed to generate 30,000 UA reads while in Run 2, 45 to 98 minutes were needed to generated 30,000 UA reads (Fig. 2A, Supplemental Fig. 1) when 5 samples were included in the same run. If the run time instead of cost is a primary concern and one flow cell is being used dedicatedly for one sample, sufficient reads can be generated for the test in less than 15 minutes (Fig. 2A). In our tests, aneuploidy detection results using 30,000 to 60,000 UA sequencing reads were concordant with results obtained using VeriSeq NGS tests for whole-chromosome aneuploidy detection (Fig. 2B, Table 1). The full trisomy and monosomy cases were detected using 30K or more UA reads and matched with the VeriSeq NGS test results (Fig. 2B, Table 1).
Large CNV Detection
The capability of performing large CNV detection using nanopore sequencing was primarily investigated using 60,000 UA reads on sample 6. Sample 6 is a female with trisomy 13, monosomy 14, and gain on chr15 73M-110M. A large CNV gain on chr 15 (70M-90M) in sample 6 were detected using 60000 UA nanopore sequencing reads. This shows the potential of performing large CNV detection using nanopore sequencing. However, future experiments are needed to systematically investigate and improve the detection sensitivity and limitation (Supplemental Fig. 2A).
Mosaicism Detection
Our nanopore-based method was primarily developed to perform rapid aneuploidy screening. However, this method can, in theory, also screen high-level mosaic aneuploidy. Current VeriSeq NGS were developed to screen-out 20% to 80% mosaic aneuploidy using 600K to 900K reads, representing 1–4 cells carrying full aneuploidy in a 5-cell biopsy. Sample 7 is a female trisomy 15, monosomy 18, and carries a 20% to 40% mosaic trisomy on chr6 as determined by VeriSeq NGS. When performing aneuploidy detection using 60,000 UA reads, the trisomy 15 and monosomy 18 were detected, and mosaic trisomy chr6 were detected (Table 1, Supplemental Fig. 2B). However, the mosaic trisomy on chr6 was not detected when using 30,000 UA reads (Table 1). Thus, it is possible to detect mosaic aneuploidy using nanopore sequencing, but it also requires longer run times (≥3 h vs. 1–2 h) to generate deeper coverage for high confidence.
Study Limitations
A 1 hour to 2-hour sequencing run generated sufficient data to perform full aneuploidy detection for up to 5 samples (including a reference control) but is not sufficient for reliable detection of large CNVs or mosaic aneuploidy detection. For that, 2-hour or longer sequencing runs may be required at current sequencing speed, or fewer samples can be run in a single flow cell. In addition, as with other NGS-based approaches, this method would not be able to detect triploid or tetraploid cases when the sex chromosomes are in the same ratio as a normal sample, unless sequencing depth was increased significantly to enable single nucleotide polymorphism-based screening. Finally, this approach also relies on uniform amplification of the whole genome as an amplification failure can result in noisy results; hence PGS detection failure.
DISCUSSION
Here we present the first successful application of nanopore sequencing technology (third generation sequencing) to preimplantation genetic screening (PGS). Our barcoded library preparation of short DNA fragments enabled rapid simultaneous copy number assessment of all chromosomes from five samples on a single, low-cost, hand-held DNA sequencer, and could be scaled by running multiple nanopore sequencers in parallel.
There are several advantages of a nanopore-based technique compared to a traditional NGS-based system. First, low initial capital costs and a small physical footprint mean that nanopore technology could be accessible to individual fertility clinics and obviate the need to transport samples to large reference laboratories which adds additional cost, time and risk for loss or damage of specimens (16, 18, 22). Multiplexing multiple samples with barcoded adapters to run five samples simultaneously reduces reagent and equipment cost per-sample to ~$150, which is comparable to the reagent cost of an Illumina VeriSeq PGS assay (~$120–$150 per sample) (18). In addition, the rapid library preparation and sequencing can facilitate same day test-and-transfer of euploid embryos and therefore avoid the need to freeze all embryos and delay transfer while waiting for PGS results, and the additional cost that incurs. In theory, nanopore-based sequencing could also allow simultaneous assessment of aneuploidy (PGS) as well as detection of a predetermined point mutation (PGD) if the WGA-amplified and targeted-amplified DNA were sequenced together, but this would need to be tested and validated.
In contrast to the second generation NGS platforms, such as those from Illumina and Ion Torrent, nanopore-based sequencing delivers sequencing results on a read-by-read basis in real time (16, 22). By reducing the number of samples run on a single nanopore flow-cell or by increasing the number of flow cells used, or sequencing run time, the number of reads on each sample could also be increased further. Increasing the number of reads obtained would allow successful detection of smaller CNVs and mosaicism. For example, to detect a 20 Mb duplication or deletion, ~ 60,000 reads would be required, based on extrapolation of our current data. Since the detection of aneuploidy is based on the number of reads aligning to each individual chromosome, the sequencing time necessary for detecting aneuploidy on individual chromosome, particularly of the larger chromosomes such as Chr1 and Chr2, would be even shorter than the time necessary to confirm that the sample is euploid on all chromosomes.
In viewing the results of this study, caution is required. This was a pilot study and was performed on a small number of samples to test feasibility, and all samples had been successfully tested by VeriSeq assay. Future prospective studies will be needed to test in larger sample size, and prior to or in parallel with current NGS- or microarray-based PGS testing in order to better estimate the test performance statistics (23). Implementing a sequencing platform in individual fertility clinics will also require regulatory oversight, training and a high degree of robustness of the platform. While we were able to theorize the ability of the assay to detect mosaicism and CNVs, additional studies will be required to evaluate these aspects of testing. Thus, in the evolution of methods to better assess chromosomal copy number in the preimplantation embryo, from FISH to reverse transcription-polymerase chain reaction, microarray and NGS (24–26), nanopore-based sequencing has the potential to offer rapid testing results within an IVF lab setting and represents an important advancement in preimplantation genetic testing.
CONCLUSION
Nanopore-based sequencing is a promising sequencing platform that can enable rapid PGS detection onsite. With a 2.5-hour standard WGA amplification from cell-biopsy, 45-minute library preparation workflow and under 2 hours for sequencing of 5 multiplexed samples, normal samples and samples with full aneuploidy were detected correctly. A single sample can be sequenced within 20 minutes and with 2-hour or longer sequencing run, detection of large CNVs and mosaic aneuploidy is possible.
Supplementary Material
Acknowledgment:
We thank Drs. Thomas Tuschl, Jan Vijg, and Yousin Suh, members of the Columbia University Fertility Center and the Williams laboratory for their helpful input and advice with this project and article; and the team at Combimatrix, in particular Mark McDonough, Karine Hovanes, Trilochan Sahoo, and Natasa Dzidic for providing the PGS DNA.
Supported by the National Institutes of Health (grants HD068546 and U19CA179564).
Footnotes
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/users/16110-fertility-and-sterility/posts/33966-25859
S.W. is listed as an inventor on patent applications covering the use of short read nanopore sequencing. Z.R.W. has nothing to disclose. P.G. has nothing to disclose. E.F. has nothing to disclose. Z.W. is listed as an inventor on patent applications covering the use of short read nanopore sequencing.
REFERENCES
- 1.Lee E, Illingworth P, Wilton L, Chambers GM. The clinical effectiveness of preimplantation genetic diagnosis for aneuploidy in all 24 chromosomes (PGD-A): systematic review. Hum Reprod 2015;30:473–83. [DOI] [PubMed] [Google Scholar]
- 2.Fragouli E, Alfarawati S, Daphnis DD, Goodall Nn, Mania A, Griffiths T, et al. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod 2011;26:480–90. [DOI] [PubMed] [Google Scholar]
- 3.Lee H-L, McCulloh DH, Hodes-Wertz B, Adler A, McCaffrey C, Grifo JA. In vitro fertilization with preimplantation genetic screening improves implantation and live birth in women age 40 through 43. J Assist Reprod Genet 2015;32:435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman EJ, Hong KH, Franasiak JM, Scott RT. Obstetrical and neonatal outcomes from the BEST Trial: single embryo transfer with aneuploidy screening improves outcomes after in vitro fertilization without compromising delivery rates. Am J Obstet Gynecol 2014;210:157.e1–6. [DOI] [PubMed] [Google Scholar]
- 5.Forman EJ, Tao X, Ferry KM, Taylor D, Treff NR, Scott RT. Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum Reprod 2012;27:1217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril 2013;100:100–7.e1. [DOI] [PubMed] [Google Scholar]
- 7.Guidance on the limits to the number of embryos to transfer: a committee opinion. Fertil Steril 2017;107:901–3. [DOI] [PubMed] [Google Scholar]
- 8.Kane SC, Willats E, Bezerra Maia e Holanda Moura S, Hyett J, da Silva Costa F. Pre-implantation genetic screening techniques: Implications for clinical prenatal diagnosis. Fetal Diagn Ther 2016;40:241–54. [DOI] [PubMed] [Google Scholar]
- 9.Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod 2013;88:69, 1–6. [DOI] [PubMed] [Google Scholar]
- 10.Fiorentino F, Biricik A, Bono S, Spizzichino L, Cotroneo E, Cottone G, et al. Development and validation of a next-generation sequencing–based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril 2014;101:1375–82.e2. [DOI] [PubMed] [Google Scholar]
- 11.Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, et al. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod 2014;29:2802–13. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Wang X, Zhang J, Song Z, Wang S, Gao Y, et al. Detection of Chromosomal Aneuploidy in Human Preimplantation Embryos by Next-Generation Sequencing1. Biol Reprod 2014;90:95, 1–6. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, Jin H, Liu L, Liu J, Wang W-H. Application of next-generation sequencing for 24-chromosome aneuploidy screening of human preimplantation embryos. Mol Cytogenet 2015;8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kung A, Munné S, Bankowski B, Coates A, Wells D. Validation of next-generation sequencing for comprehensive chromosome screening of embryos. Reprod Biomed Online 2015;31:760–9. [DOI] [PubMed] [Google Scholar]
- 15.Ruttanajit T, Chanchamroen S, Cram DS, Sawakwongpra K, Suksalak W, Leng X, et al. Detection and quantitation of chromosomal mosaicism in human blastocysts using copy number variation sequencing. Prenat Diagn 2016;36:154–62. [DOI] [PubMed] [Google Scholar]
- 16.Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 2016;17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei S, Williams Z. Rapid Short-Read Sequencing and Aneuploidy Detection Using MinION Nanopore Technology. Genetics 2016;202:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei S, Weiss ZR, Williams Z. Rapid Multiplex Small DNA Sequencing on the MinION Nanopore Sequencing Platform. G3 (Bethesda) 2018;8:1649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17:10. [Google Scholar]
- 20.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res 2002;12:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafaye de Micheaux P, Drouilhet R, Liquet B. The R Software. New York, NY: Springer New York; 2013. [Google Scholar]
- 22.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC genomics 2012;13:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Niu W, Peng Z, Bao X, Zhang M, Wang L, et al. Comparative study of single-nucleotide polymorphism array and next generation sequencing based strategies on triploid identification in preimplantation genetic diagnosis and screen. Oncotarget 2016;5:81839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srebniak MI, Knapen MFCM, Polak M, Joosten M, Diderich KEM, Govaerts LCP, et al. The influence of SNP-based chromosomal microarray and NIPT on the diagnostic yield in 10,000 fetuses with and without fetal ultrasound anomalies. Hum Mutat 2017:1–20. [DOI] [PubMed] [Google Scholar]
- 25.Carlson LM. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet Gynecol Clin North Am 2017;44:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handyside AH. 24-chromosome copy number analysis: a comparison of available technologies. Fertil Steril 2013;100:595–602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


