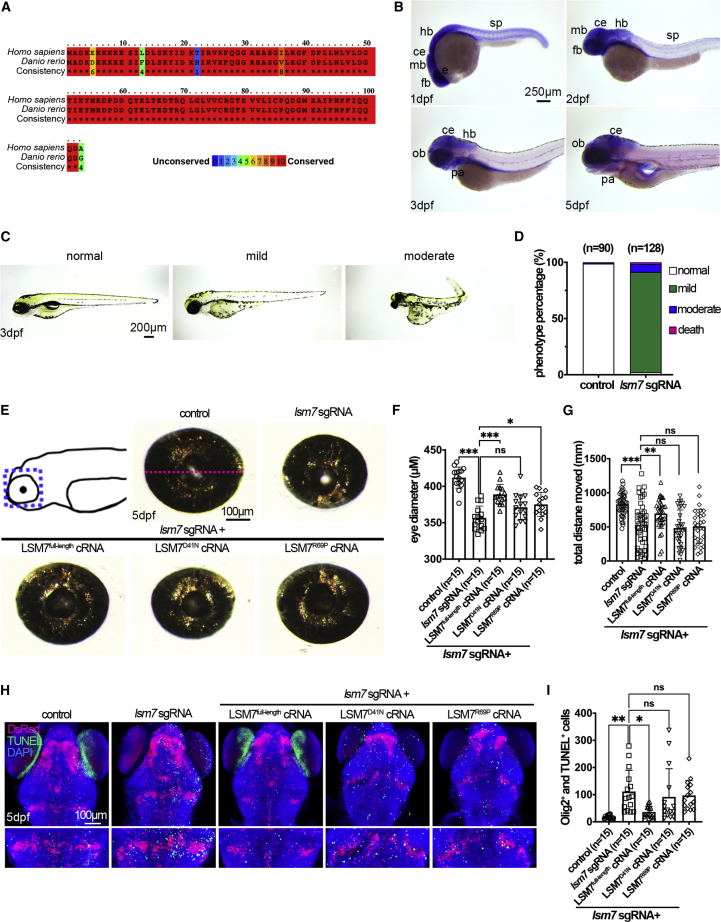
Figure 5.
The zebrafish Lsm7 ortholog is highly conserved with human LSM7, is expressed in developing nervous system, and is necessary for embryonic morphology, eye size, motor behavior, and survival of oligodendrocytes
(A) Amino acid alignment of human and zebrafish LSM7 sequences. Identical residues are marked by red boxes, and percent similarity is shown by different colors.
(B) In situ hybridization was performed for detecting lsm7 expression during zebrafish embryogenesis. Embryo stages are shown in the bottom left of each panel. All panels display the lateral view, with dorsal to the top and rostral to the left.
(C) Phenotypes of lsm7-Crispants.
(D) Quantification of control and lsm7-Crispants by phenotype severity.
(E) Top left panel shows a cartoon representation of the dissected region of larvae (blue dashed line rectangle). Images of eyes of control, lsm7-Crispant, LSM7full-length cRNA-, LSM7R69P cRNA-, and LSM7D41N cRNA-rescued larvae are shown. The eye diameter was measured as indicated by the magenta dashed line shown on control eye.
(F) Quantification of eye size.
(G) Motor swimming behavior of the control, lsm7-Crispant, LSM7full-length cRNA-, LSM7R69P cRNA-, and LSM7D41N cRNA-rescued larvae at 7 dpf.
(H) Confocal images of dorsal view of the control, lsm7-Crispant, LSM7full-length cRNA-, LSM7R69P cRNA-, and LSM7D41N cRNA-rescued larvae with rostral to the top. Lower panels are enlarged region from upper panels for apoptotic cell quantification.
(I) Quantification of the number of TUNEL and DsRed double-positive cells of (H). Abbreviations: ce, cerebellum; dpf, days post-fertilization; fb, forebrain; hb, hindbrain; mb, midbrain; ob, olfactory bulb; pa, pharyngeal arches; sp, spinal cord. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., not significant.