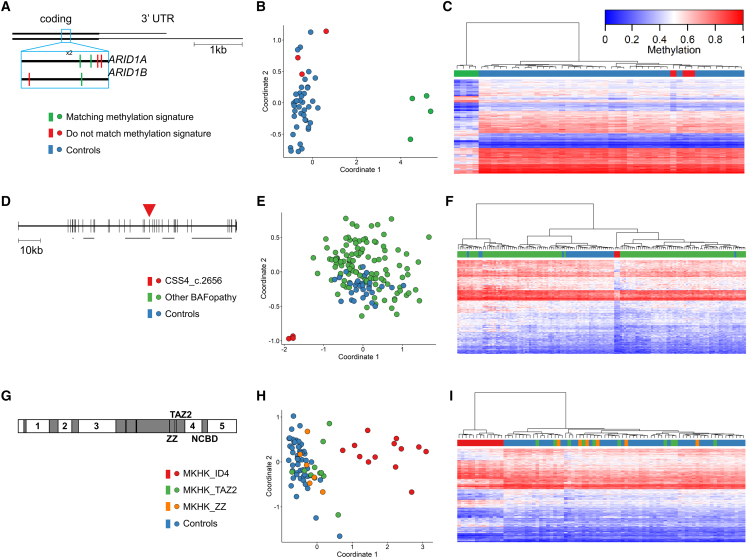
Figure 2.
Gene region- or variant-specific sub-signatures
(A) The last exon of ARID1A and ARID1B shown with the location of seven variants in the c.6200 region colored by whether they match the c.6200 episignature or not. (B and C) MDS (B) and hierarchical clustering (C) plots of the seven samples showing that the four central samples have a matching episignature, while the outer three cluster with controls. For hierarchical clustering plots, each row represents one microarray probe, and each column represents one sample. (D) Gene diagram of SMARCA4 (NM_001128849.1) showing the location of the three c.2656A>G variants in exon 19 (red arrowhead). The five horizontal gray bars indicate the locations of protein domains: QLQ, HSA, helicase ATP-binding, helicase C-terminal, and bromodomain. (E and F) MDS (E) and hierarchical clustering (F) showing that the three CSS4 samples with the above variant cluster separately from controls and from other BAFopathy samples. (G) Protein diagram of CREBBP/EP300 showing the location of protein domains (gray boxes) and intrinsically disordered (ID) domains (numbered). (H and I) MDS (H) and hierarchical clustering (I) showing the MKHK_ID4 samples clustering separately from controls and from other MKHK samples.