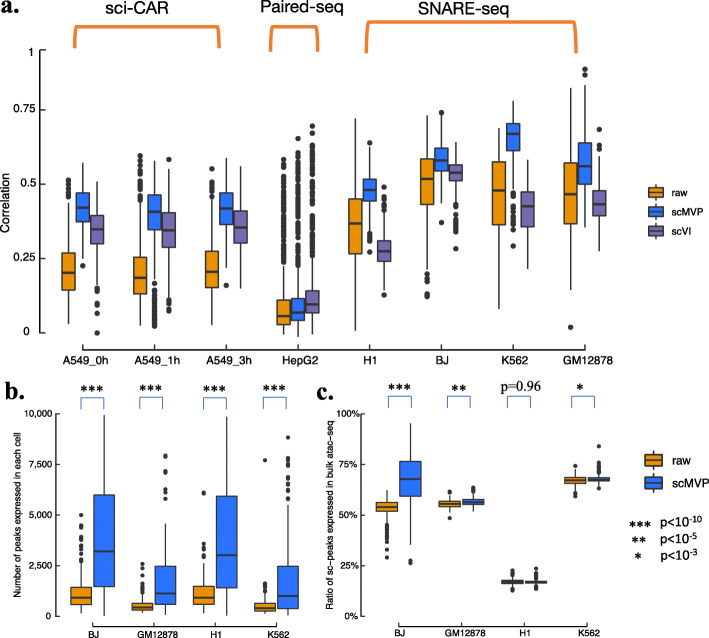
Fig. 2.
scMVP mitigates data sparsity in joint profiling datasets. a Correlation between original and imputed gene expression of each cell from scRNA of joint profiling datasets and gene expression in corresponding bulk RNA-seq dataset. A549 cell lines treated with DEX for 0h (ENCSR632DQP), 1h (ENCSR656FIH), 3h (ENCSR624RID) in sci-CAR dataset, HepG2 cell line (ENCSR058OSL) in Paired-seq dataset and H1 (ENCSR670WQY), BJ (ENCSR000COP), K562 (ENCSR530NHO), and GM12878 (ENCSR000CPO) cell lines in SNARE-seq dataset were used for benchmark. b Number of bulk ATAC peaks identified by raw and imputed scATAC in each cell. c Ratio of raw and imputed scATAC peaks identified in bulk ATAC peaks. DNase-seq signal files for H1 (ENCSR000EMU) and BJ (ENCSR000EME), and ATAC-seq signal files for K562 (ENCSR868FGK) and GM12878 (ENCSR095QNB) were used for benchmark