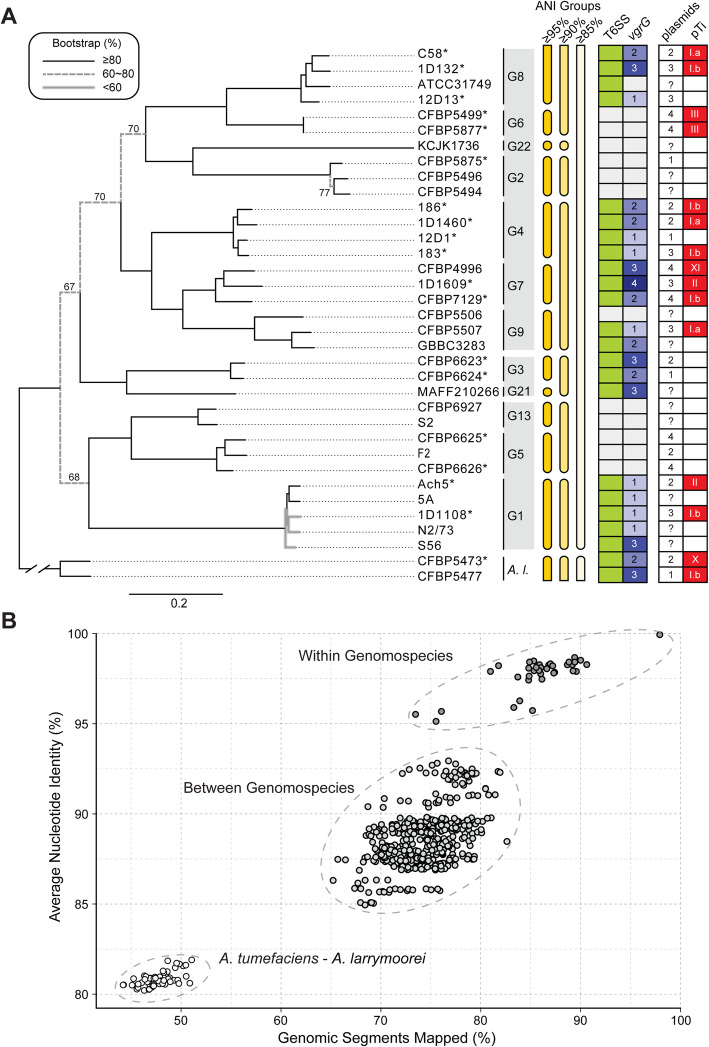
Fig. 1.
Relationships among representatives of the Agrobacterium tumefaciens species complex. The sister species Agrobacterium larrymoorei (A. l.) is included as the outgroup. A Maximum likelihood phylogeny based on a concatenated alignment of 2093 single-copy genes shared by all 35 genomes (635,594 aligned amino acid sites). Bootstrap support values in the range of 60–80% are labeled. Strains with complete genome assemblies are highlighted with an asterisk (“*”). The genomospecies assignments (i.e., G1-G9, G13, and G21-G22) are labeled to the right of strain names. The three A. tumefaciens supergroups are indicated by the colored background of the genomospecies assignments. Information to the right of the genomospecies assignments shows the grouping of genomes according to different cutoff values of genome-wide average nucleotide identity (ANI), the presence/absence of type VI secretion system (T6SS)-encoding gene cluster (green: present; white: absent), copy number of vgrG (white background: absent), number of plasmids, and the tumor-inducing plasmid (pTi) type based on k-mer profile (white background: absent). B Pairwise genome similarities based on the percentages of genomic segments mapped and the ANI values