Figure 6.
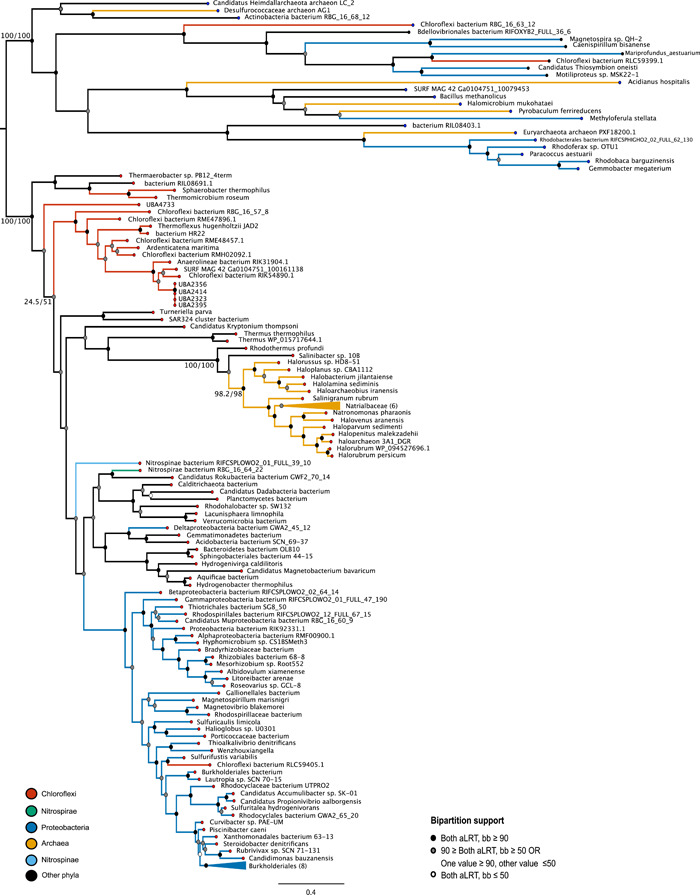
eNOR gene tree. A phylogenetic tree of homologs to the nitric oxide reductase from SURF MAG 42 reveals an expanded diversity of putative eNOR subunit I homologs in not only Archaea and Chloroflexi, but also Proteobacteria and other diverse phyla. Putative eNOR sequences (red tips) have the characteristic Gln‐323 in the alignment; outgroup sequences (blue tips) have Tyr‐323 (oxygen reductase superfamily) or other substitutions. Support values for selected bipartitions are labeled (aLRT/bb). Support for other nodes is indicated with the following color scheme: Strong support with both values ≥90 (black); weak support with both values ≤50 (white); intermediate support with one or both values between 50 and 90 (gray); conflicting support, with one value ≤50 and the other ≥90 (gray). MAG, metagenome‐assembled genome; SURF, Sanford Underground Research Facility
