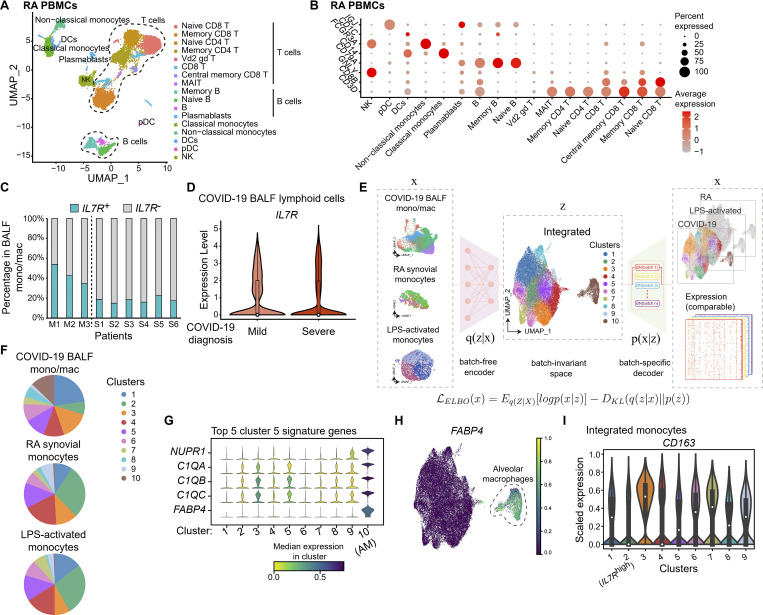
Figure S5.
CD127 expression designates a functionally distinct monocyte subset. (A) UMAP projection of PBMCs from an RA patient. Cell type annotations were labeled for each cluster. DC, dendritic cell; pDC, plasmacytoid dendritic cell. (B) Heat map shows the expression of hallmark genes in different cell clusters from RA PBMCs. The scaled average expression levels of marker genes and the percentage of cells expressing marker genes are expressed by color and size of each dot corresponding to cell clusters, respectively. MAIT, mucosal associated invariant T cell; NK, natural killer cell. (C) The percentages of IL7R+ cells in BALF monocytes/macrophages (mono/mac) from mild or severe COVID-19 patients are individually shown for each patient. (D) Lymphoid cells from COVID-19 patient BALF were subgrouped by the diagnosed disease severity. Violin plot shows the expression of IL7R in BALF lymphoid cells from mild or severe COVID-19 patients. Each overlaid box indicates the interquartile range with the median shown as a circle. (E) The schematic plot briefly shows the methodology of SCALEX. The asymmetric variational autoencoder (VAE) framework was structured for SCALEX via a batch-free encoder and a batch-specific decoder, in which batch-free encoder extracted the batch-invariant biological features (z) that were masked by batch-related variations among the input scRNA-seq datasets (x), and batch-specific decoder incorporated the batch information when reconstructing the expression matrix for integrated data. Such a probabilistic model of SCALEX is shown in the panel and is briefly introduced in Materials and methods. On this basis, we integrated monocytes from COVID-19 BALF, RA synovial tissue, and LPS-treated monocytes by SCALEX to generate a comparable integrated scRNA-seq dataset with preservation of the biological features for each incorporated component. (F) Pie graphs projecting the percentage of each cluster in E in total monocytes or macrophages from different tissue sources. (G) The stacked violin plot shows the expression of the top five cluster 10 signature genes in all 10 clusters shown in E. Median expression levels for each gene in each cluster were indicated by colors. (H) UMAP projection of integrated monocytes/macrophages in E. FABP4 expression in cells was quantitatively visualized by the indicated colors. Cells corresponding to cluster 10 were highlighted by a dotted line as alveolar macrophages, given the specific FABP4 expression pattern. (I) Violin plot shows the expression of CD163 among nine clusters of integrated monocytes in Fig. 6 G, in which cluster 2 was designated as the IL7Rhigh cluster. Each overlaid box indicates the interquartile range with the median shown as a circle.