Figure 1.
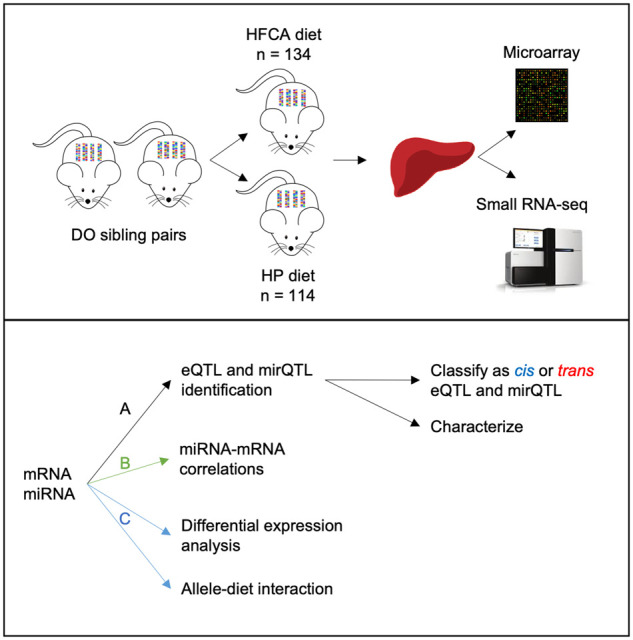
Experimental and analytical design classifies, characterizes, and explores the relationships between hepatic mRNA and miRNA in the context of diet. A total of 292 female DO mice were obtained from Jackson Laboratory as 146 full sibling pairs. After initial maintenance on a synthetic diet (AIN-76A), one sibling from each pair was randomly assigned either an HFCA or HP diet for 18 weeks. Extracted hepatic mRNA was hybridized to the Affymetrix Mouse Gene array and miRNA was sequenced via Illumina HiSeq. mRNA and miRNA expression values were used as phenotypes in four QTL genome scans. (A) The four models were diet as an additive covariate (n = 243), diet as an interactive covariate (n = 243), subset of HFCA-fed mice (n = 134), and subset of HP-fed mice (n = 109). eQTL and mirQTL found in each model were classified as cis or trans and characterized. (B) Interactions between miRNA and mRNA were assessed and compared between models. (C) The role of diet was explicitly analyzed by differential expression analysis and by identifying eQTL with significant allele-diet interactions from regression analysis.
