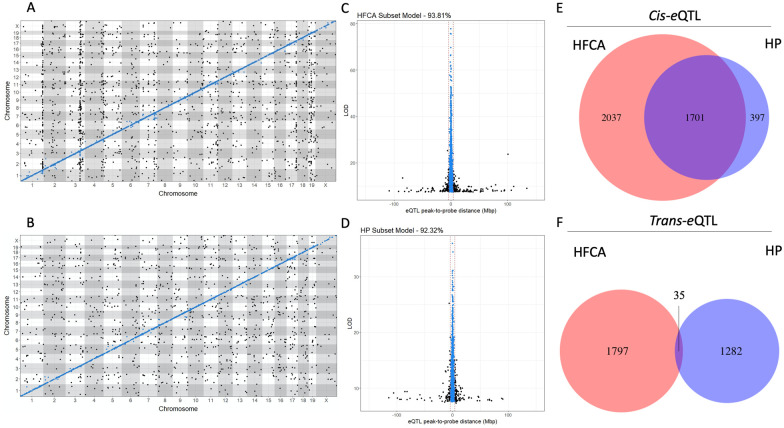
Figure 7.
Analysis of genome scans from HFCA- and HP-fed mice reveal environmentally driven differences in genetic architecture. (A) Global eQTL architecture for the HFCA diet model shows numerous, dense trans-bands, in contrast to the (B) HP diet model, which shows notably less colocalization of trans-eQTL. Both diet models (C and D) show similar precisions from eQTL with probe sets on the same chromosome; resolution of these 4036 eQTL from HFCA-fed mice and 3648 eQTL from HP-fed mice are estimated to be 0.355 and 0.359 Mb, with 88.8 and 89.2% of cis-eQTL occurring ±4 Mb from their probe sets, respectively. This indicates that structural variants associated with local genetic regulation tend to occur close to the gene itself, regardless of environmental perturbations. (E, F) A large number of cis-eQTL overlap despite environmental differences, while the lack of overlap in the trans-eQTL might be indicative of environmental effects.