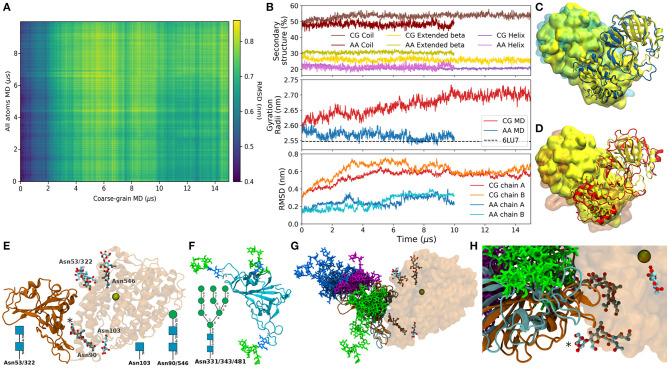
Figure 2.
(A) Bi-dimensional RMSD of the Cα between the atomic trajectory and the backmapped conformations from the CG trajectory. (B) Top: Percentage of secondary structure content. Middle: Gyration Radii of the whole proteins from the atomic (blue), and backmapped trajectory in red. Bottom: RMSD of the Cα between the trajectories and the experimental structure, separated by chains. (C) Superposition between the experimental structure (yellow) and the last conformation of the atomic trajectory (blue). Chains A are presented as cartoon and chains B as presented as surface. (D) Same as (C) with the backmapped structure of the last frame of the SIRAH trajectory in red. (E) X-ray structure PDB 6VW1 in cartoon representation, ACE2 is semitransparent, glycosylations are show as sticks colored according to element. The Zinc ion present in the binding site is shown as a space-filling sphere. The asterisk marks the glycosylation at Asn90 on ACE2. The N-glycosylation solved by X-ray on each position are indicated schematically. (F) Starting conformer of the RBD glycosylated at Asn331, 343, and 481 colored according to the SNFG color scheme (46). (G) Superposition of backmapped structures from the glycosylated RBD and that in the structure 6VW1. Proteins are colored according to panels (E) and (F). RBD Glycosylations sites are shown in magenta, blue, and green for Asn331, Asn343, and Asn481, respectively. Only one conformed every 1 μs is shown. (H) Close up on the RBD-ACE2 interaction showing the positions of Asn481 (green), and the closest glycans in ACE2.