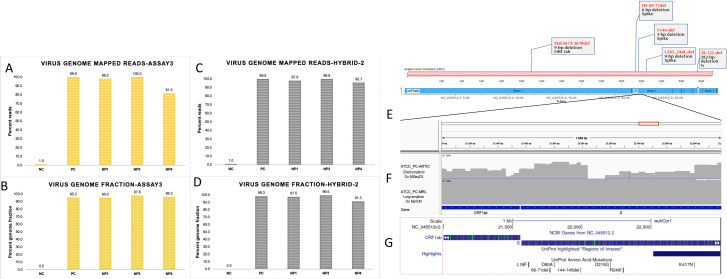
Fig 6. Long-read sequencing provides uniform coverage across deletion-prone region in the virus genome.
Panel A shows the percentage of the virus genome mapped sequencing reads (A) and virus genome fraction covered with Nanopore sequencing data (B), in the positive control (VR1986D ATCC SARS-CoV-2 RNA). Panels C-D show reads mapped to the virus genome and covered genomic fraction in combined, ARTIC + Long-read (Hybrid-II) data, respectively. Panel E illustrates a gene sketch on known deletions in 2kb region of the spike gene. Panel F shows sequencing coverage across a deletion-prone region of the spike gene in ATCC positive control RNA in short-amplicon ARTIC and long-amplicon MRL data. Top coverage plot on ATCC_PC_ARTIC shows sequencing coverage in short-amplicon on Illumina MiSeqDx platform, whereas second track underneath show long-read sequencing data on ATCC_PC sample using MRL long-amplicon primers. Panel G shows a UCSC genome browser snapshot across spike gene region that show previously reported deletions in this genomic interval of SARS-CoV-2 genome.