Abstract
Facing the unstopped surges of COVID-19, an insufficient capacity of diagnostic testing jeopardizes the control of disease spread. Due to a centralized setting and a long turnaround, real-time reverse transcription polymerase chain reaction (real-time RT-PCR), the gold standard of viral detection, has fallen short in timely reflecting the epidemic status quo during an urgent outbreak. As such, a rapid screening tool is necessitated to help contain the spread of COVID-19 amid the countries where the vaccine implementations have not been widely deployed. In this work, we propose a saliva-based COVID-19 antigen test using the electrical double layer (EDL)-gated field-effect transistor-based biosensor (BioFET). The detection of SARS-CoV-2 nucleocapsid (N) protein is validated with limits of detection (LoDs) of 0.34 ng/mL (7.44 pM) and 0.14 ng/mL (2.96 pM) in 1× PBS and artificial saliva, respectively. The specificity is inspected with types of antigens, exhibiting low cross-reactivity among MERS-CoV, Influenza A virus, and Influenza B virus. This portable system is embedded with Bluetooth communication and user-friendly interfaces that are fully compatible with digital health, feasibly leading to an on-site turnaround, an effective management, and a proactive response taken by medical providers and frontline health workers.
Keywords: Covid-19, SARS-CoV-2, Electrical double layer, Field-effect transistor-based biosensors, Rapid antigen tests
1. Introduction
As the new hotspots were hit by the unstopped surges of COVID-19 [1], the reported cases have surpassed 208 million worldwide as of August 2021 [2]. COVID-19, an ongoing pandemic with fast-evolving variants, is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerging as the most impactful threat to global health in a century [3]. The cumulative death toll has reached over 4.3 million since the outbreak was declared by the World Health Organization (WHO) in 2020 [2]. Early symptoms of COVID-19 are similar to a common flu-like illness; yet in serious cases, patients may suffer dyspnea and proceed with severe pneumonia, acute respiratory distress, multiple organ dysfunction, septic shock, etc. [4]. Vaccination, which reduces the risk of severe COVID-19 [5], [6], is regarded as the most effective tool against the viral transmission; whereas the treatments remain unclear and mostly rely on supportive care [7], [8]. As such, rapid detection, effective management, and proactive responses are necessitated to contain the spread of COVID-19 across the countries where vaccine implementations have not been widely deployed.
COVID-19 diagnostics can be sorted into two categories [9]: viral tests (also known as diagnostic test) and antibody tests. Viral tests, such as molecular tests (for viral RNA) and antigen tests (for viral protein), diagnose active infection of patients; while antibody tests are serological tests reflecting past infection [10]. The real-time reverse transcription polymerase chain reaction (real-time RT-PCR), the gold standard for SARS-CoV-2 viral tests, is an in vitro diagnostics (IVDs) where a sample is usually collected through a nasal swab [11], [12]. This nucleic acid-based testing can detect as low as ~100 copies/mL of the viral RNA [13], but its sensitivity varies from 70% (real-world tests) to 99% (an ideal condition) [14], [15], [16], [17]. The turnaround time of a real-time RT-PCR test usually takes from 4 h to 2 days, and it needs to be operated by highly skilled personnel in a centralized lab [18].
Several rapid antigen testing techniques were approved of Emergency Use Authorizations (EUAs) by the U.S. Food and Drug Administration (FDA) [19], [20]. A lateral flow immunochromatographic assay (LFIA) provides a qualitative detection for COVID-19 [21], [22], while a chemiluminescence enzyme immunoassay (CLEIA) offers a quantitative measurement of SARS-CoV-2 antigens [23]. Compared to PCR-based techniques, the testing time of a viral antigen detection is tremendously reduced (within 60 min) [16], [22]. However, the sensitivity is usually compromised (60 – 80%) [22], [23], and the semi-invasive specimen collection using nasal, nasopharyngeal, or oropharyngeal swaps brings discomfort to testees. As such, a salivary detection, which avails a noninvasive sample collection, has been considered as an alternative method for rapid COVID-19 screenings. Moreover, viral loads found in saliva, ranging from 104 copies/mL to 108 copies/mL, are comparable with what are found in nasal cavities and throats [24], [25], [26], [27], [28], [29], [30]. Amongst novel antigen tests developed for COVID-19 [16], [22], [23], [31], field-effect transistor-based biosensors (BioFET) are of significant advantages as per a high sensitivity, a wide dynamic range, a real-time readout, and a matrix-insensitivity across a wide variety of analytes [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41]. Nanomaterial-based BioFETs demonstrate the excellent candidacy for low-concentration measurements [31], [35], [38]. BioFETs using high electron mobility transistors (HEMTs) are utilized to detect miRNA [37], peptide [33], [39], SARS-CoV-1 nucleocapsid (N) protein [34], circulating tumor cells (CTCs) [40], etc. Though the reported BioFETs using nanomaterials [31], [32], [35], [38], [41] or HEMTs [33], [34], [37], [39], [40] are highly sensitive, their costs, reusability, and portability must be improved before deploying for in situ COVID-19 immunoassays. As such, a portable BioFET featuring low cost, disposable testing sticks, good sensitivity, and salivary detection should be developed to address the needs for on-site COVID-19 screenings.
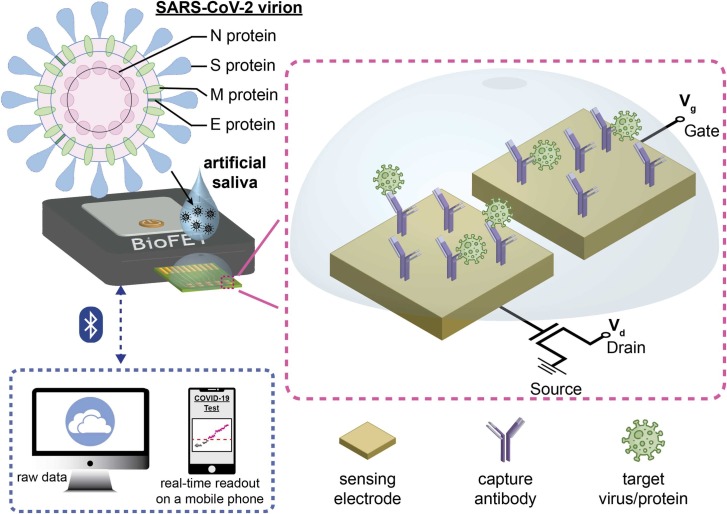
In this work, we developed a saliva-based antigen test of SARS-CoV-2 N protein using an electrical double layer (EDL)-gated BioFET system ( Fig. 1). The proposed system included a portable reader functioned with Bluetooth where a testing result can be immediately displayed on a smartphone using mobile-based user interface (UI). The ease of pretreatment and the digital health-compatible setting enabled a fast turnaround time (within 30 min). EDLs were redistributed along with reactions on surfaces, and the changes in EDL capacitance allowed BioFETs to detect analytes in a variety of physiological conditions (e.g., serum, blood, saliva, etc.) [32], [42]. Surface functionalization was verified with fluorescence imaging, and sensor-to-sensor variation is discussed. The COVID-19 antigen tests using EDL-gated BioFETs were validated in both 1× PBS and artificial saliva, and the limits of detection (LoDs) were calculated. To investigate cross-reactivity, the antigens of MERS-CoV, Influenza A virus, and Influenza B virus were tested. Aiming to find a diagnostic niche, the antigen tests in artificial saliva using an EDL-gated BioFET can progress toward the detection of clinical samples (human saliva). This rapid testing can timely reflect the epidemic status quo (e.g., the number of infected individuals) and benefit the policymaking, fighting against the spread of COVID-19.
Fig. 1.
Schematic illustration of a saliva-based COVID-19 antigen test using an electrical double layer (EDL)-gated field-effect transistor biosensor (BioFET). An artificial slaiva sample consisting of SARS-CoV N portein is drop-casted on a sensor stick, and a testing result is displayed on a smart phone via Bluetooth in 30 min.
2. Materials and methods
2.1. The BioFET system for COVID-19 viral antigen tests
The custom-designed BioFET platform, as shown in Fig. 1 and Supplemental Fig. 1, consisted of a disposable sensor stick, a portable reader (CC&C Technologies, Taiwan) embedded with a Bluetooth function, and two custom-written UIs operated for Microsoft Windows and iOS, respectively. Each sensor stick (Jumpers Biotech, Taiwan), which was custom-designed and fab-manufactured, had 8 individually addressable sensors arranged in an 1 × 8 array where each sensor comprised of two gold electrodes (500 × 500 µm2) on a 75-µm pitch. SU-8 photoresist (Kayaku Advanced Materials, #SU8–2010) was coated on a sensor stick, and an active area (450 × 450 µm2) of each electrode was photolithographically defined. An input gate voltage (Vg) was applied on one of the electrodes (of each sensor), and an output Vg was measured at the gate terminal of an FET via the other electrode (of each sensor). The Bluetooth-embedded reader transmitted data to the devices where a real-time result was displayed on an iPhone, and raw data were stored in a laptop for further analysis.
2.2. Surface functionalization
A sensor stick was placed in an O2 plasma cleaner (Harrick Scientific Products, USA, #PDC-32G) for 180 s at a constant power of 18 W (high RF level), then the sensor stick was rinsed with 10% HCl (Sigma-Aldrich, #320331) and DI water, successively. The anti-SARS-CoV-2 N protein antibody (GeneTex, Taiwan, #GTX632269), simply named “anti-N antibody” throughout the rest of content, was used as the capture antibody. 14 mM of Traut’s Reagent (Thermo Fisher Scientific, #26101) was dissolved in PBS-EDTA (1× PBS, with 5 mM of EDTA) prior to mixing with 1.5 mg/mL of anti-N antibody (volume ratio = 1:10) at room temperature for 1 h. 11 μL of thiolated antibody, formed through the previous procedure, was detached from an excess amount of Traut’s Reagent using a desalting column (Thermo Fisher Scientific, #89877) which was equilibrated with PBS-EDTA. The thiolated antibody was diluted with PBS-EDTA at a volume ratio of 1 : 1, and the final concentration was 0.68 mg/mL. 0.5 μL of diluted antibody solution was then drop-casted on each sensor where the immobilization took place at 14 – 18 °C for 12 h. Finally, the functionalized sensors were rinsed with 1 mL of 1× PBS to remove the unbound antibody.
2.3. Fluorescence imaging
Anti-Mouse IgG (GeneTex, Taiwan, #GTX213111–05), the secondary antibody bound to the capture antibody (anti-N antibody), was labeled with a fluorescent dye (DyLight 594). 50 μL of the solution, in the presence of fluorophore-labeled antibody (2 µg/mL), was drop-casted on a sensor stick (covering all the eight sensors) and incubated at room temperature for 1 h. Afterwards, the sensor stick was rinsed with 1 mL of 1× PBS and the unbound fluorophore-labeled antibody was removed. An optical measurement was taken by a fluorescence microscope (Leica Microsystems, #DM2500 LED) where a result was analyzed and quantified using Leica LAS X and Image J.
2.4. Proteins and immunoassays
In PBS-based immunoassays, the desired concentrations of SARS-CoV-2 N protein (GeneTex, Taiwan, #GTX135357-pro), SARS-CoV-2 S protein (Leadgene Biomedical, Taiwan, #61831), MERS-CoV N protein (GeneTex, Taiwan, #GTX135653-pro), Influenza A virus nucleoprotein (GeneTex, Taiwan, #GTX135868-pro), and Influenza B virus nucleoprotein (GeneTex, Taiwan, #GTX135867-pro) were respectively spiked into 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4 at pH = 7.4 with NaOH). In saliva-based immunoassays, 100 μM of sodium dodecyl sulfate (SDS) (Thermo Fisher Scientific, #28312) was dissolved in clinically mimetic matrix where artificial saliva (Pickering Laboratories, USA, #1700–0305) was mixed with the universal transport medium (UTM) (COPAN Diagnostics, USA, #330 C) at a volume ratio of 1:1. The mixture of artificial saliva, SDS, and UTM, is simply named as “artificial saliva” throughout the rest of content. Following the same procedures, the desired concentrations of SARS-CoV-2 N protein, SARS-CoV-2 S protein, MERS-CoV N protein, Influenza A virus nucleoprotein, and Influenza B virus nucleoprotein were respectively spiked into artificial saliva. Both kinds of immunoassays were performed in the presence of capture probes (anti-N antibody) which were immobilized on a sensor surface. 70 μL of a testing solution was drop-casted on a sensing area, and signals were measured/recorded using the custom-designed BioFET platform.
2.5. FET characteristics and signal acquisition
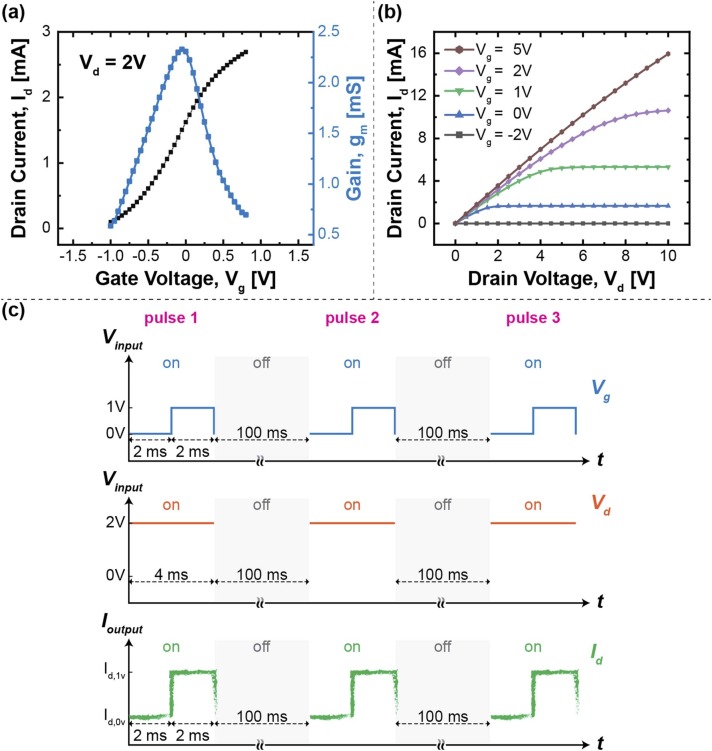
N-channel depletion-mode DMOS FETs (Microchip Technology, #LND150) (n = 8) were electrically characterized by a semiconductor parameter analyzer (Agilent, #B1500A) prior to mounting on a printed circuit board (PCB) (Supplemental Fig. 1). The transfer characteristics of the FET are shown in Fig. 2a, the maximum transconductance takes place near Vg = 0 V at a constant source-drain voltage (Vd) of 2 V. The FET characteristics of drain current (Id) versus Vd are displayed in Fig. 2b.
Fig. 2.
(a) Transfer characteristics, and (b) Id-Vd characteristics at different gate biases of a FET. (c) Signal acquisition. The inputs were applied with a constant Vd and three pulses of Vg during each measurement, while the output signals (Ich) were retrieved by the difference between two current levels.
The COVID-19 antigen tests were taken at a constant Vd (2 V) with a square wave of gate biases (Vg = 0 V for 2 ms followed by Vg = 1 V for 2 ms) as shown in Fig. 2c. The elapsed time of each measurement was set as 212 ms where three pulses of Vg were applied discretely with two intermediate turnoffs. The output Id was measured at a sampling rate of 167 kHz, and Ich was the characteristic current at which the difference between two current levels was calculated:
| (1) |
| (2) |
| (3) |
where is the averaged Id calculated within the last 1 ms of the nth pulse at Vg = 0 V, and is the averaged Id calculated within the last 1 ms of the n th pulse at Vg = 1 V.
3. Results and discussion
The BioFET platform adopted the outreach configuration, where gate terminals of the FETs were extended via wires and connected to a sensor stick, to prevent direct corrosion of a testing sample on FETs. To overcome the Debye screening while enabling detection in a physiological condition (e.g., serum, blood, saliva, etc.), EDL-gated BioFETs were leveraged to measure double-layer capacitance rather than surface charges. As such, sample pretreatment can be tremendously eased, and a turnaround time is significantly reduced (<1 h) [32], [37], [39], [40], [42]. The detailed sensing mechanism using an EDL-gated BioFET can be found in Supplemental Fig. 2.
To amplify an electrical signal, the FETs measured a testing sample at a linear region (Vg = 1 V) and a saturation region (Vg = 0 V), respectively (as described in Materials and Methods). A high Vd causes a heating effect that gives rise to a noisy background and a signal drift, yet a low Vd yields a small transconductance. As a trade-off, Vd was set as 2 V to achieve a higher conductance (compared to Vd = 1 V) while producing a minor heating and an acceptable noise. The data were retrieved and collected every 2 min, and totally 11 measurements (20 min) were taken for each concentration of analytes.
3.1. Surface functionalization
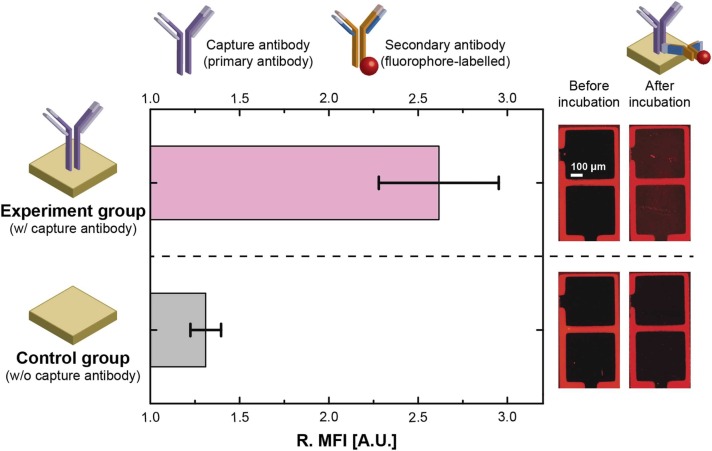
To confirm successful surface functionalization, a fluorescent measurement was performed. A sensor stick was split into two groups: three (out of eight) sensors were treated with buffer solution, serving as the control group; while the other five sensors were functionalized with capture antibody, serving as the experiment group. After incubation of fluorophore-labeled secondary antibody, the optical tags (i.e., fluorophores) were excited at 593 nm and emitted red fluorescence at 618 nm. A mean fluorescence intensity (MFI) was quantified within a quarter of an electrode using ImageJ, and 8 subareas were measured for a sensor. The background induced 14.08 ± 0.05 A.U. of MFI prior to incubation of secondary antibody as shown in Supplemental Fig. 3. In the control group, a minor amount of the secondary antibody remained on the surface after the washing step, emitting 18.49 ± 1.16 A.U. of MFI. While the experiment group exhibited at least 28.40 A.U. of MFI (Supplemental Fig. 3). The representative images of an unfunctionalized sensor (S#1) and a functionalized sensor (S#4) are shown in Fig. 3, and the brightness indicates the amount of the fluorophore-labeled secondary antibody. The relative MFI (R. MFI) was defined as the ratio of an MFI measured after incubation of secondary antibody to an MFI measured before incubation of secondary antibody (). Error bars represent one standard deviation (1σ) of uncertainties measured across sensors as shown in Fig. 3 (n = 5 in the experiment group, n = 3 in the control group). The experiment group exhibited 2× the R.MFI of the control group, indicating a successful functionalization that can be employed for the succeeding immunoassays. While the sensor-to-sensor variation of R. MFI can be attributed to nonuniform immobilization of capture antibody.
Fig. 3.
Optical quantification of surface funcctionalization (left) and fluorescent images (right). The relative mean fluorescence intensity (R. MFI) was calculated by the MFI measured before/after incubation of secondary antibody. The control group exhibits 2.62 A.U. of R. MFI. Error bars represent 1σ of sensor-to-sensor uncertainty measured by fluoroscence intensity.
3.2. Saliva-based COVID-19 antigen tests using BioFETs
The structural proteins of SARS-CoV-2 are majorly composed of envelope (E) protein, transmembrane (M) protein, N protein, and spike (S) protein. N protein is abundantly expressed during an infection, thus it is regarded as a highly immunogenic protein and was selected for the antigen tests in this work [43]. To investigate the sensor response to SARS-CoV-2 viral protein; the desired concentrations of SARS-CoV-2 N protein, ranging from 0.4 ng/mL to 400 ng/mL, were prepared in 1× PBS. The testing samples were drop-casted onto a sensor stick successively varying from lowest to highest concentration, and electrical measurements were taken every two minutes using BioFETs. Prior testing sample was removed from the sensor stick before the next testing sample was added. A baseline of each series of measurement was defined as where a norm measurement was first taken in the absence of an analyte (N protein), and the subsequent BioFET signals were measured with a subtracted baseline:
| (4) |
where is the measured at [N protein] = j ng/mL, and is the measured at [N protein] = 0 ng/mL. BioFET measurements usually took several minutes to get signal stabilized after spiking analytes (due to temperature drift, diffusion, binding kinetics, etc.), so 8 out of 11 measurements were used to calculate a mean signal at each concentration.
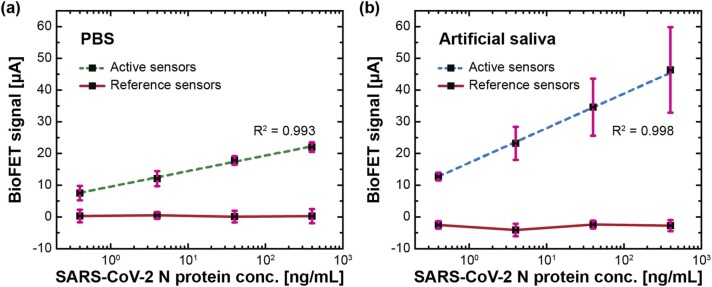
In the controlled experiment, reference sensors were tested in the absence of an immobilized antibody (anti-N antibody), and the increasing concentrations of viral N protein had an unremarkable effect on a sensor response (variation < 3 µA) as shown in Fig. 4a. This indicates that non-specific binding was negligible. While the active sensors, immobilized with capture antibody, linearly responded to the added SARS-CoV-2 N protein (in a logarithmic scale) that the concentrations ranged from 0.4 ng/mL to 400 ng/mL. The sensor-to-sensor variation, as shown in Supplemental Fig. 4, may result from a nonuniform coverage of capture antibody (Fig. 3). To benchmark the sensor performance and quantify a LoD, the method of the Clinical and Laboratory Standards Institute (CLSI) was adopted [44], [45]:
| (5) |
| (6) |
where LoB is the limit of blank, is the standard deviation of the result measured from the low concentration sample, is the mean result of the blank sample, and is the standard deviation of the result measured from the blank sample. The overall change in signal was 22.0 µA, and the calculated LoD was 342.16 pg/mL (7.44 pM).
Fig. 4.
COVID-19 antigen tests using EDL-gated BioFETs in (a) 1× PBS and (b) artificial saliva. Active sensors were functionalized with capture antibody, while reference sensors were unfunctionalized. SARS-CoV-2 N protein concentration varied from 0.4, 4, 40, to 400 ng/mL. Error bars represent ± 1σ of uncertainty measured by sensors (n = 3).
To validate COVID-19 antigen tests using BioFETs in a more realistic scenario, the measurements were taken in artificial saliva (as described in Materials and Methods). Saliva is viscous and tends to congeal quickly after collection, making it difficult to be pipetted for further liquid-based measurements. As such, UTM was used to mix with artificial saliva due to its stability at room temperature when collecting as well as transporting viral samples [46], [47]. Plus, the detergent (SDS), which can break a coat of the enveloped virus by denaturing a viral membrane or causing a conformational change, was added [48]. Following the same procedure of the PBS-based immunoassay, only the medium was replaced with artificial saliva. The reference sensors exhibited signal variations less than 5 µA (Fig. 4b), and the use of artificial saliva induced an opposite change in capacitance compared to what was measured in 1× PBS. In the experiment group, the overall change in signal was 46.33 μA with a good linearity (R2 = 0.998), and the calculated LoD was 136.25 pg/mL (2.96 pM). The change of matrices exhibited comparable LoDs, and an addition of detergent (SDS) did not interfere with the assay. While an one-fold increase in the overall signals might be a consequence of inhomogeneous surface functionalizations and different media. The real-time results of COVID-19 antigen tests, in both 1× PBS and artificial saliva, can be found in Supplemental Fig. 5. Considering a small volume (500 nL per sensor) used in surface functionalization, experimental uncertainty (e.g., manual pipetting) led to inhomogeneous surface coverages. In addition, biological complexity in artificial saliva brought on electrical fluctuation more formidably than in PBS. Overall, the testing time of each concentration was 20 min, and the turnaround time was less than 30 min, effectuating rapid COVID-19 antigen tests using an EDL-gated BioFET.
3.3. Investigation of cross-reactivity
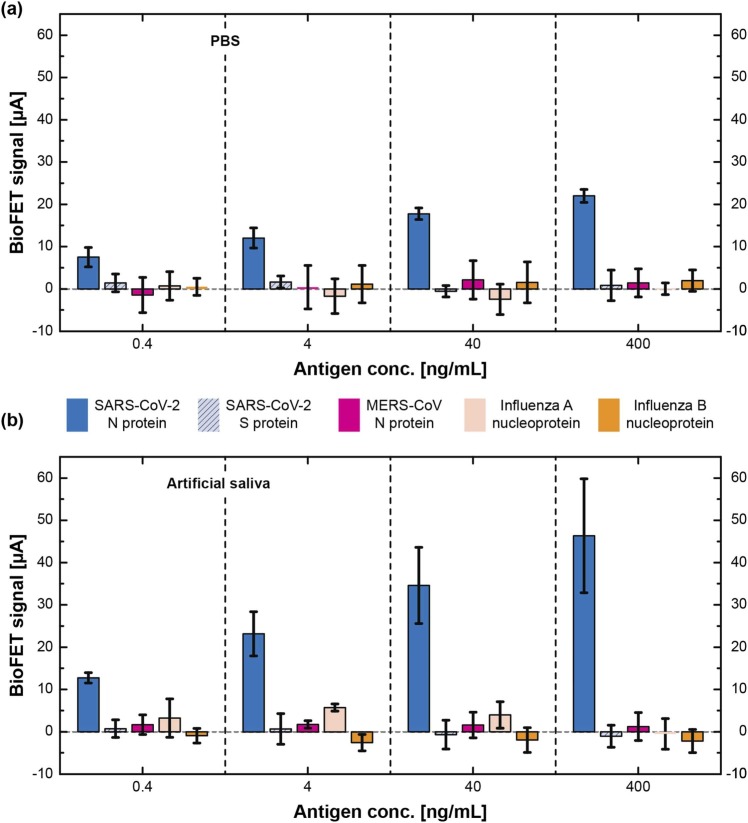
To further inspect the specificity, various antigens were tested with EDL-gated BioFETs. SARS-CoV-2 S protein, MERS-CoV N protein, Influenza A virus nucleoprotein, and Influenza B virus nucleoprotein were spiked into artificial saliva, drop-casted on a sensor stick where its sensor surfaces were functionalized with anti-N antibody. The data of SARS-CoV-2 N protein shown in Fig. 5 are extracted from Fig. 4, enabling a visual comparison of the cross-reactivity. Among the groups of the lowest testing concentration (0.4 ng/mL), the specificities are relatively insignificant in both matrices. Notably, the signals of different antigens (except SARS-CoV-2 N protein) were located within the variations which were 3 uA for PBS and 5 uA for artificial saliva, respectively; and no increasing/decreasing trend was found in all cases. The detection specificities improved as the concentrations of antigens increased (>4 ng/mL), and SARS-CoV-2 N protein eventually achieved 11.21× the signal of other antigens at a concentration of 400 ng/mL in PBS. While the measurements in artificial saliva yielded signal-to-cross-reactivity ratios () of 4.04, 8.73, and 21.03 at concentrations of 4 ng/mL, 40 ng/mL, and 400 ng/mL, respectively. Though the higher signals were found in artificial saliva, the detection uncertainty was more significant in artificial saliva than PBS. This phenomenon can be attributed to the extra electrolytes and chemicals which may complicate the molecular environment. Taken together, EDL-gated BioFETs demonstrated good specificities (signal-to-cross-reactivity ratio > 4.04) in both PBS and artificial saliva when antigen concentrations were higher than 4 ng/mL, indicating a negligible cross-reactivity.
Fig. 5.
Investigation of cross-reactivity in (a) PBS and (b) artificial saliva. The data of SARS-CoV-2 N protein are retrieved from Fig. 4 and are replotted here for the comparison of cross-reactivity. Error bars represent ± 1σ of uncertainty measured by sensors (n = 3).
3.4. Testing landscape and comparison of COVID-19 diagnostics
As of August 2021, low vaccination rates and insufficient capacity of diagnostic testing have fueled the new cases of COVID-19 worldwide. To fight against the spread of COVID-19, a critical solution is to field diagnostic tools which have a high accuracy, a fast turnaround, a portable configuration, an user-friendly operation/readout delivering quantitative results, and a digital health-compatible setting [16], [49]. Several proposed tools have received EUA [19], [20], [50], yet the governmental action primarily relies on the reported cases confirmed by real-time RT-PCR. Ideally a turnaround time of real-time RT-PCR requires couples of hours ( Table 1) [12], [50], however, the delayed deliveries of samples/results between infrastructures induce the issue of testing backlogs [51]. Due to centralized testing, a limited capacity, and excessive numbers of samples during an outbreak, sole reliance on PCR-based results have conceivably hampered the policymaking against the spread of COVID-19, leading to misjudgment of the epidemic status quo and obscureness of the disease control [51].
Table 1.
Comparison of COVID-19 diagnostics.
| Molecular tests | Antibody tests | Antigen tests | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Target | RNA (RdRp, E gene, N gene) | RNA (ORF-1a, E gene) | RNA (N gene, S gene) | N Ab | N Ab, S Ab (S1, S1S2) | S Ab | N Protein | N protein | S protein (S1) | N protein, S protein (S1) | N protein |
| Testing specimen | NPS, NS | NPS, NS, OPS | NPS | Serum | Serum | Human plasma | NS | NPS, NS | PBS, 0.01x UTM, culture medium, NPS | DPBS, saliva | Artificial saliva |
| Dilution | – | – | – | No | 1:1600 in PBST | 1:1000 in PBS | – | – | – | 1:2 in DPBS | 1:1 with UTM |
| Methodology | Real-time RT- PCR |
Real-time RT- PCR |
Electro-chemical biosensor | SPR | GC-FP | LSPR | LFIA | CLEIA | Graphene-based BioFET |
Glucometer (electrochemical biosensor) | EDL-gated BioFET |
| Portability | No, centralized | No, centralized | Yes, handheld | Yes, hand-carried | No, centralized | No, centralized | Yes, handheld | No, centralized | No, centralized | Yes, handheld | Yes, handheld |
| Size (mm3) | – | – | 157 × 97 × 35 | 175 × 155 × 55 | – | – | – | – | – | 76 × 48 × 16 | 120 × 80 × 30 |
| Commercial Availability | Off-the-shelf device | Off-the-shelf device | Off-the-shelf device + lab-engineered testing strips | Off-the-shelf device + lab-engineered testing chips | Lab prototype | Lab prototype | Off-the-shelf device | Off-the-shelf device | Lab prototype | Off-the-shelf device + lab-engineered testing strips | Lab prototype |
| Highlights | – | – | Isothermal RCA | – | – | – | – | – | – | Aptamer-based competitive assay | Bluetooth-embedded, mobile-based UI |
| Testing time | 2 hr | 3 – 8 h | N/A | 15 min | 30 min | 30 min | – | – | 1 – 2 min | < 5 min | 2 – 20 min |
| Turnaround time | > 4 hr | 1 day | 2 hr | 60 – 90 min | ~1 hr | ~1 hr | 15 min | 2 – 4 hr | < 1 hr | ~65 min | 30 min |
| Quantification | Yes | No | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes |
| LoD* | 3.6 – 3.9 copy/rxn | 1.8 × 103 ndu/mL | 1 copy/μL | 1 µg/mL | < 2 ng/spot | ~0.5 pM | 1.58 × 102 TCID50/mL | 2.2 × 101 TCID50/mL | 13.1 aM (PBS)a 1.31 fM (UTM)a 16 pfu/mL (CM)a 242 copies/mL (CS)a |
DPBS: 1.50 pM (N protein)b 1.31 pM (S protein)b Saliva: 5.27 pM (N protein)b 6.31 pM (S protein)b |
7.44 pM (PBS)c 2.96 pM (AS)c |
| Reference | [12] | [52] | [53] | [54] | [55] | [56] | [22] | [23] | [31] | [16] | This Work |
Ab: antibody. AS: artificial saliva. CM: culture medium. CS: clinical sample. DPBS: Dulbecco’s potassium phosphate buffered saline. GC-FP: grating-coupled fluorescent plasmonics.
LSPR: localized surface plasmon resonance. NPS: nasopharyngeal swab. NS: nasal swab. OPS: oropharyngeal swab. PBST: phosphate buffered saline with Tween-20. RCA: rolling circle amplification.
RdRp: RNA-dependent RNA polymerase. SPR: surface plasmon resonance. TCID50: median tissue culture infectious dose.
* The methods of defining LoDs: a the lowest concentration detected by a sensor; b the slope method where ; and c the CLSI method, please refer to the main text in this article.
To address the needs, some diagnostic tests (e.g., molecular tests and antigen tests) and antibody tests using commercially-available devices and/or lab prototypes have been proposed as shown in Table 1 [12], [16], [21], [22], [23], [31], [50], [52], [53], [54], [55], [56]. In general, molecular tests exhibit the best sensitivity/specificity, yet centralized settings and slow turnarounds deteriorate disease control. Chaibun et al. developed a portable electrochemical biosensor for molecular tests, while the LoD was not as low as the conventional PCR-based methods are [13], [53]. Several antibody tests using surface plasmon-based techniques, which have an intermediate turnaround, were developed to verify a past infection; whereas the LoDs were traded off against the complexity of pretreatments and the portability of a device [54], [55], [56]. Amongst novel methods developed for antigen testing, Seo et al. detected SARS-CoV-2 S protein using graphene-based BioFETs that was ultrasensitive and provided the LoDs (242 copies/mL in clinical samples) comparable to PCR-based methods, yet this nano device had to be measured using a bulky semiconductor analyzer in a centralized lab [31]. Singh et al. utilized an off-the-shelf glucometer with custom-engineered test strips to validate COVID-19 antigen detection in human saliva, and the LoDs reached in the range of few pM, yielding high accuracy of 100% of positive percent agreement (PPA) (n = 16) and 100% of negative percent agreement (NPA) (n = 8) in clinical testing [16]. While the 4-step pretreatment using aptamers and magnetic beads prolonged the turnaround time to ~65 min. To find a diagnostic niche, we developed a saliva-based COVID-19 antigen test using an EDL-gated BioFET system. Considering its LoD (~3 pM), a diagnosis of active infection, a quantitative result, a compatibility to a digital health using Bluetooth communication and mobile-based UI, a handheld portability (120 × 80 × 30 mm3), and a fast turnaround (30 min); the proposed system detecting SARS-CoV-2 N protein in artificial saliva owns a high potential to be deployed amid the frontline of diagnostic screening.
4. Conclusion
Endeavoring to fight against COVID-19, we successfully developed an antigen test of SARS-CoV-2 N protein in artificial saliva using an EDL-gated BioFET system. This portable system can be fielded for on-site COVID-19 screening since the matrix insensitivity simplifies a pretreatment and the digital health-compatible setting eases a data outputting/collection, speeding up a turnaround time to 30 min. Surface functionalization was verified with fluorescence imaging, and sensor-to-sensor variation could root in a nonuniform coverage of surface functionalization. The detections of SARS-CoV-2 N protein were corroborated in 1× PBS and artificial saliva, indicating LoDs of 342.16 pg/mL (7.44 pM) and 136.25 pg/mL (2.96 pM), respectively. The cross-reactivity was minor, and specificity increased as the antigen concentration exceeded 4 ng/mL. The proposed system validated COVID-19 antigen tests in artificial saliva, while the assessment of clinical samples and deployments around medical infrastructures will be processed when receiving the approval/authorization from the Institutional Review Board (IRB). The testing of clinical samples collected from human saliva is expected to be more challenging since human saliva consists of extra electrolytes, enzymes, proteins, cells, mucus, etc., increasing the complexity of detections.
CRediT authorship contribution statement
Pin-Hsuan Chen: Conceptualization, Methodology, Software, Investigation, Writing – original draft, Visualization. Chih-Cheng Huang: Methodology, Validation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Chia-Che Wu: Methodology, Investigation. Po-Hsuan Chen: Investigation. Adarsh Tripathi: Writing – review & editing. Yu-Lin Wang: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported, in part, by research grants from Ministry of Science & Technology, Taiwan (MOST 109–2218-E-007–017), SPARK Program, Taiwan (109Q2901E1) and National Tsing Hua University, Taiwan (109Q2805E1, 109Q2706E1). We thank the technical support from National Nano Device Laboratories (NDL) in Hsinchu and the Center for Nanotechnology, Materials science, and Microsystems (CNMM) at National Tsing Hua University.
Biographies

Pin-Hsuan Chen received the B.S. degree in biomedical engineering and environmental sciences from National Tsing Hua University, Hsinchu, Taiwan, in 2019, and the M.S. degree in power mechanical engineering from National Tsing Hua University, Hsinchu, Taiwan, in 2021. Her principal research interests are electrochemistry and biosensors. She worked on the development of a rapid screening biosensor platform that detected SARS-COV-2 virus.

Dr. Chih-Cheng Huang received the B.S. degree in materials science and engineering from National Taiwan University, Taipei, Taiwan, in 2010, the M.S. degree in nanoengineering from National Tsing Hua University, Hsinchu, Taiwan, in 2012, and the Ph.D. degree in materials science from the University of California San Diego, La Jolla, CA, USA, in 2020. He is currently a postdoctoral research fellow at National Tsing Hua University. His research interests include nanomagnetism, PoC diagnostics, next-generation biosensing techniques, and proteomics. Dr. Huang has been a member of the American Chemical Society (ACS) since 2012.

Chia-Che Wu received the B.S. degree in biological science and technology from China Medical University, Taichung, Taiwan in 2018, and the M.S. degree in pharmacology from National Taiwan University, Taipei, Taiwan, in 2020. He is currently a master student at National Tsing Hua University. His research focuses on electrochemical biosensing and its application on early diagnoses.

Dr. Po-Hsuan Chen received the B.S. degree in biological science and technology from China Medical University, Taichung, Taiwan in 2009, the Ph.D. degree in molecular biology from National Chung Cheng University, Chia-Yi, Taiwan, in 2016. His research interests include molecular biology, cancer biology, cell biology, biochemistry, and semiconductor-based bio-sensors.

Adarsh Tripathi received his Integrated Masters degree (5 years integrated program) in Biotechnology from Vellore Institute of Technology, Vellore, Tamil Nadu, India in 2014. He was a Ph.D. student in National Tsing Hua University. His research interests include genetic engineering, proteomics and biosensing.

Dr. Yu-Lin Wang received his B.S. degree in chemistry from Tunghai University and M.S. degree from National Taiwan University, in 1993 and 1995, respectively. He had worked in semiconductor industry from 1997 to 2006. He received his Ph.D. in materials science and engineering from University of Florida, in 2009. He is currently a Professor in the Institute of NanoEngineering and Microsystems, Department of Power Mechanical Engineering, at National Tsing Hua University, Hsinchu, Taiwan. His research interests are semiconductor-based sensors and the device for medical use and personal healthcare. His team has won several awards in recent years including the Top 10 pioneering technology worldwide selected by Google-X in 2016, Merit of Asia Pacific ICT Aliance in 2016, Gold Medal by Spintech Inc. in 2018, Silver Medal by uTAS in 2018, and Silver Medal by EpiStar Inc. in 2019.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.131415.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.S.P. and L. Magalhaes, South America Is Now Covid-19 Hot Spot, With Eight Times the World’s Death Rate, Wall Str. J. (2021). https://www.wsj.com/articles/south-america-is-now-covid-19-hot-spot-witheighttimes-the-worlds-death-rate-11624299176 (Accessed August 19, 2021).
- 2.World Health Organization, Weekly epidemiological update on COVID-19 - 17 August 2021, 2021. 〈https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---17-august-2021〉 (Accessed August 19, 2021).
- 3.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Coronaviridae study group of the international committee on taxonomy of viruses, the species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 5.Hippisley-Cox J., Coupland C.A., Mehta N., Keogh R.H., Diaz-Ordaz K., Khunti K., Lyons R.A., Kee F., Sheikh A., Rahman S., Valabhji J., Harrison E.M., Sellen P., Haq N., Semple M.G., Johnson P.W.M., Hayward A., Nguyen-Van-Tam J.S. Risk prediction of covid-19 related death and hospital admission in adults after covid-19 vaccination: national prospective cohort study. BMJ. 2021;374:n2244. doi: 10.1136/bmj.n2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson M.G., Stenehjem E., Grannis S., Ball S.W., Naleway A.L., Ong T.C., DeSilva M.B., Natarajan K., Bozio C.H., Lewis N., Dascomb K., Dixon B.E., Birch R.J., Irving S.A., Rao S., Kharbanda E., Han J., Reynolds S., Goddard K., Grisel N., Fadel W.F., Levy M.E., Ferdinands J., Fireman B., Arndorfer J., Valvi N.R., Rowley E.A., Patel P., Zerbo O., Griggs E.P., Porter R.M., Demarco M., Blanton L., Steffens A., Zhuang Y., Olson N., Barron M., Shifflett P., Schrag S.J., Verani J.R., Fry A., Gaglani M., Azziz-Baumgartner E., Klein N.P. Effectiveness of Covid-19 Vaccines in Ambulatory and Inpatient Care Settings. N. Engl. J. Med. 2021;385:1355–1371. doi: 10.1056/NEJMoa2110362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 8.Mallapaty S. Can COVID vaccines stop transmission? Scientists race to find answers. Nature. 2021 doi: 10.1038/d41586-021-00450-z. [DOI] [PubMed] [Google Scholar]
- 9.United States Centers for Disease Control and Prevention COVID-19 testing overview. Cent. Dis. Control Prev. 2020 〈https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/testing.html〉 accessed June 6, 2021. [Google Scholar]
- 10.United States Food and Drug Administration, Coronavirus Disease 2019 Testing Basics, FDA, 2021. 〈https://www.fda.gov/consumers/consumer-updates/coronavirus-disease-2019-testing-basics〉 (Accessed July 5, 2021).
- 11.Xiao A.T., Tong Y.X., Gao C., Zhu L., Zhang Y.J., Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnaout R., Lee R.A., Lee G.R., Callahan C., Cheng A., Yen C.F., Smith K.P., Arora R., Kirby J.E. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., Campo R.D., Ciapponi A., Sued O., Martinez-García L., Rutjes A.W., Low N., Bossuyt P.M., Perez-Molina J.A., Zamora J. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. ELife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021;180 doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Yang M., Yuan J., Wang F., Wang Z., Li J., Zhang M., Xing L., Wei J., Peng L., Wong G., Zheng H., Wu W., Shen C., Liao M., Feng K., Li J., Yang Q., Zhao J., Liu L., Liu Y. Laboratory diagnosis and monitoring the viral shedding of SARS-CoV-2 infection. Innovation. 2020;1 doi: 10.1016/j.xinn.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei X., Lee H.-C., Diao K., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.United States Food and Drug Administration, In Vitro Diagnostics EUAs, (2021). 〈https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas〉 (Accessed July 5, 2021).
- 20.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.United States Food and Drug Administration, Individual EUAs for Antigen Diagnostic Tests for SARS-CoV-2, Vitro Diagn. EUAs - Antigen Diagn. Tests SARS-CoV-2. (2021). https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-antigen-diagnostic-tests-sars-cov-2 (Accessed July 26, 2021).
- 22.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M.Á., Martínez M., Poujois S., Forqué L., Valdivia A., Solano de la Asunción C., Ferrer J., Colomina J., Navarro D. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag rapid test device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021;27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefever S., Indevuyst C., Cuypers L., Dewaele K., Yin N., Cotton F., Padalko E., Oyaert M., Descy J., Cavalier E., Van Ranst M., André E., Lagrou K., Vermeersch P. Comparison of the quantitative diasorin liaison antigen test to reverse transcription-PCR for the diagnosis of COVID-19 in symptomatic and asymptomatic outpatients. J. Clin. Microbiol. 2021;59:e00374–21. doi: 10.1128/JCM.00374-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fakheran O., Dehghannejad M., Khademi A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect. Dis. Poverty. 2020;9:100. doi: 10.1186/s40249-020-00728-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Tan C., Zeng J., Luo C., Hu S., Peng Y., Li W., Xie Z., Ling Y., Zhang X., Deng E., Xu H., Wang J., Xie Y., Zhou Y., Zhang W., Guo Y., Liu Z. Analysis of viral load in different specimen types and serum antibody levels of COVID-19 patients. J. Transl. Med. 2021;19:30. doi: 10.1186/s12967-020-02693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng V.C.C., Wong S.-C., Chen J.H.K., Yip C.C.Y., Chuang V.W.M., Tsang O.T.Y., Sridhar S., Chan J.F.W., Ho P.-L., Yuen K.-Y. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020;41:493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J., Guo J., Xu Y., Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J. Infect. 2020;81:e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid Detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 32.Chen K.-I., Li B.-R., Chen Y.-T. Silicon nanowire field-effect transistor-based biosensors for biomedical diagnosis and cellular recording investigation. Nano Today. 2011;6:131–154. doi: 10.1016/j.nantod.2011.02.001. [DOI] [Google Scholar]
- 33.Huang C.-C., Lee G.-Y., Chyi J.-I., Cheng H.-T., Hsu C.-P., Hsu Y.-R., Hsu C.-H., Huang Y.-F., Sun Y.-C., Chen C.-C., Li S.-S., Andrew Yeh J., Yao D.-J., Ren F., Wang Y.-L. AlGaN/GaN high electron mobility transistors for protein–peptide binding affinity study. Biosens. Bioelectron. 2013;41:717–722. doi: 10.1016/j.bios.2012.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu Y.-R., Kang Y.-W., Fang J.-Y., Lee G.-Y., Chyi J.-I., Chang C., Huang C.-C., Hsu C.-P., Huang T., Huang Y.-F., Sun Y.-C., Hsu C.-H., Chen C.-C., Li S.-S., Yeh J.A., Yao D.-J., Ren F., Wang Y.-L. Investigation of C-terminal domain of SARS nucleocapsid protein–duplex DNA interaction using transistors and binding-site models. Sens. Actuators B Chem. 2014;193:334–339. doi: 10.1016/j.snb.2013.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maehashi K., Ohno Y., Matsumoto K. Utilizing research into electrical double layers as a basis for the development of label-free biosensors based on nanomaterial transistors. Nanobiosens. Dis. Diagn. 2015;5:1–13. doi: 10.2147/NDD.S40316. [DOI] [Google Scholar]
- 36.Sakata T., Matsuse Y. In situ electrical monitoring of cancer cells invading vascular endothelial cells with semiconductor-based biosensor. Genes Cells. 2017;22:203–209. doi: 10.1111/gtc.12473. [DOI] [PubMed] [Google Scholar]
- 37.Cheng H.-L., Fu C.-Y., Kuo W.-C., Chen Y.-W., Chen Y.-S., Lee Y.-M., Li K.-H., Chen C., Ma H.-P., Huang P.-C., Wang Y.-L., Lee G.-B. Detecting miRNA biomarkers from extracellular vesicles for cardiovascular disease with a microfluidic system. Lab Chip. 2018;18:2917–2925. doi: 10.1039/c8lc00386f. [DOI] [PubMed] [Google Scholar]
- 38.Li Q., Lu N., Wang L., Fan C. Advances in nanowire transistor-based biosensors. Small Methods. 2018;2 doi: 10.1002/smtd.201700263. [DOI] [Google Scholar]
- 39.Tai T.-Y., Sinha A., Sarangadharan I., Pulikkathodi A.K., Wang S.-L., Lee G.-Y., Chyi J.-I., Shiesh S.-C., Lee G.-B., Wang Y.-L. Design and demonstration of tunable amplified sensitivity of algan/gan high electron mobility transistor (hemt)-based biosensors in human serum. Anal. Chem. 2019;91:5953–5960. doi: 10.1021/acs.analchem.9b00353. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y.-H., Pulikkathodi A.K., Ma Y.-D., Wang Y.-L., Lee G.-B. A microfluidic platform integrated with field-effect transistors for enumeration of circulating tumor cells. Lab Chip. 2019;19:618–625. doi: 10.1039/C8LC01072B. [DOI] [PubMed] [Google Scholar]
- 41.Sung D., Koo J. A review of BioFET’s basic principles and materials for biomedical applications. Biomed. Eng. Lett. 2021;11:85–96. doi: 10.1007/s13534-021-00187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu C.-H., Sarangadharan I., Regmi A., Chen Y.-W., Hsu C.-P., Chang W.-H., Lee G.-Y., Chyi J.-I., Chen C.-C., Shiesh S.-C., Lee G.-B., Wang Y.-L. Beyond the debye length in high ionic strength solution: direct protein detection with field-effect transistors (FETs) in human serum. Sci. Rep. 2017;7:5256. doi: 10.1038/s41598-017-05426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021;54:159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008;29:S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 45.Pierson-Perry J.F., Vaks J.E., Durham A.P., Fischer C., Gutenbrunner C., Hillyard D., Kondratovich M.V., Ladwig P., Middleberg R.A. Second ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2012. EP17A2 Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures.〈https://clsi.org/standards/products/method-evaluation/documents/ep17/〉 [Google Scholar]
- 46.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin. Microbiol. Infect. 2021;27:285.e1–285.e4. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berenger B.M., Conly J.M., Fonseca K., Hu J., Louie T., Schneider A.R., Singh T., Stokes W., Ward L., Zelyas N. Saliva collected in universal transport media is an effective, simple and high-volume amenable method to detect SARS-CoV-2. Clin. Microbiol. Infect. 2021;27:656–657. doi: 10.1016/j.cmi.2020.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Sousa A.L.M., Pinheiro R.R., Araújo J.F., de Azevedo D.A.A., Peixoto R.M., Andrioli A., da Cruz Silva Bezerra S.T., da Silva Teixeira M.F. Sodium dodecyl sulfate as a viral inactivator and future perspectives in the control of small ruminant lentiviruses. Arq. Inst. Biol. 2019;86 doi: 10.1590/1808-1657000752018. [DOI] [Google Scholar]
- 49.Tromberg B.J., Schwetz T.A., Pérez-Stable E.J., Hodes R.J., Woychik R.P., Bright R.A., Fleurence R.L., Collins F.S. Rapid scaling up of Covid-19 diagnostic testing in the united states — the NIH RADx initiative. N. Engl. J. Med. 2020 doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.United States Food and Drug Administration, In Vitro Diagnostics EUAs - Molecular Diagnostic Tests for SARS-CoV-2, FDA. (2021). 〈https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2〉 (accessed July 31, 2021).
- 51.Wu H. Assoc. Press News; 2021. Taiwan struggles with testing backlog amid largest outbreak.〈https://apnews.com/article/taiwan-coronavirus-pandemic-business-health-c12fc8276820e18751d85e52dcdba882〉 accessed July 31, 2021. [Google Scholar]
- 52.cobas® SARS-CoV-2, 2021. 〈https://www.fda.gov/media/136049/download〉.
- 53.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O’Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Djaileb A., Charron B., Jodaylami M.H., Thibault V., Coutu J., Stevenson K., Forest S., Live L.S., Boudreau D., Pelletier J.N., Masson J.-F. A rapid and quantitative serum test for SARS-CoV-2 antibodies with portable surface plasmon resonance sensing. ChemRxiv Camb. Camb. Open Engag. 2020;2020 doi: 10.26434/chemrxiv.12118914.v1. [DOI] [Google Scholar]
- 55.Cady N.C., Tokranova N., Minor A., Nikvand N., Strle K., Lee W.T., Page W., Guignon E., Pilar A., Gibson G.N. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Funari R., Chu K.-Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material