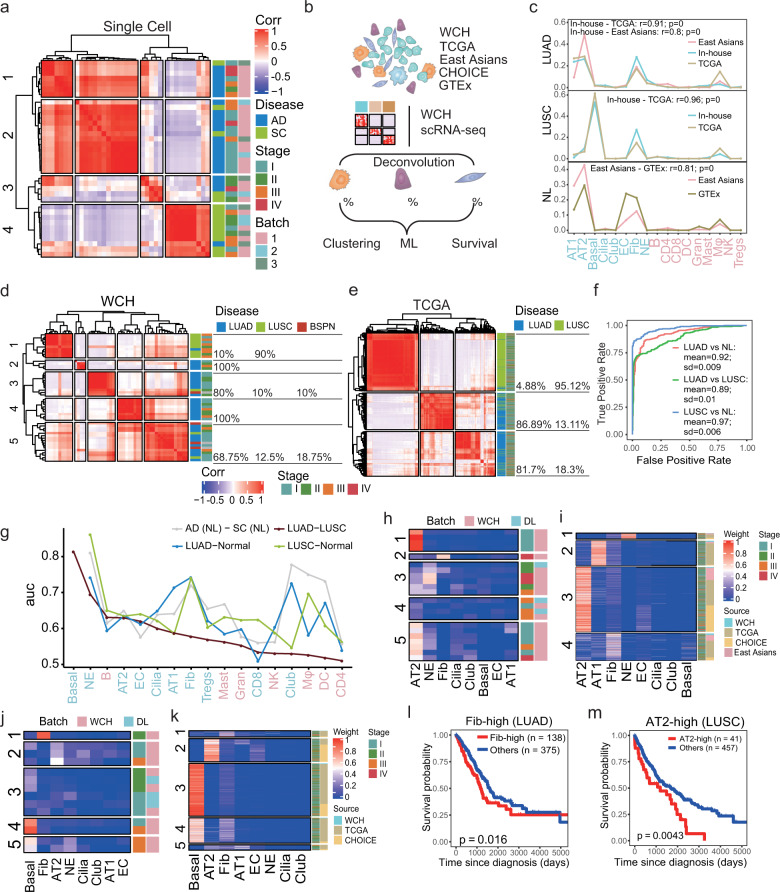
Fig. 3.
Distinct cellular composition modules of LUAD and LUSC. a Pearson correlation between cell compositions of malignant cells in scRNA-seq. b Flowchart overview of the deconvolution workflow in which our scRNA-seq was used to provide cell-type-specific genes. c Line plots show the cell composition of 17 cells deconvoluted from different sources of bulk RNA-seq. The correlations were calculated using Pearson correlation. d, e The Pearson correlation between cell weights of samples from WCH (d) and TCGA (e). LUAD and LUSC tended to be clustered together. The proportions of different disease types are labeled on the right. f ROC was performed for cell weights of different lung cancer subtypes and normal lung tissue from independent cohorts. Multiple cross-validations were performed to generate reliable values for different groups. The mean area under the curve (AUC) of LUAD vs normal tissue was 0.92 (SD = 0.009), LUSC vs normal tissue was 0.97 (SD = 0.006); LUAD vs LUSC was 0.89 (SD = 0.01). g Prioritizing the most affected cell types in LUAD and LUSC progression by ranking the AUC scores derived from the Augur algorithm. h Heatmap of the proportion of non-immune cells in patients from LUAD. i Heatmap of the deconvoluted weights of non-immune cells based on bulk RNA-seq data from LUAD patients. Patients could be separated into four groups, including NE-high (1), AT1-high (2), AT2-high (3), and Fib-high (4). j Heatmap of the proportion of non-immune cells in patients from LUSC. k Heatmap of the deconvoluted weights of non-immune cells based on bulk RNA-seq data from LUSC patients. Five groups, including Fib-high (1), AT2-high (2), Basal-high (3), Basal-Fib hybrid (4), and Hybrid (5). l Kaplan–Meier survival curves for patients with LUAD (n = 513), stratified for the Fib-high group and the rest. P value was calculated using the log-rank test. m Kaplan–Meier survival curves for patients with LUSC (n = 498), stratified for the AT2-high group and the rest. P value was calculated using the log-rank test