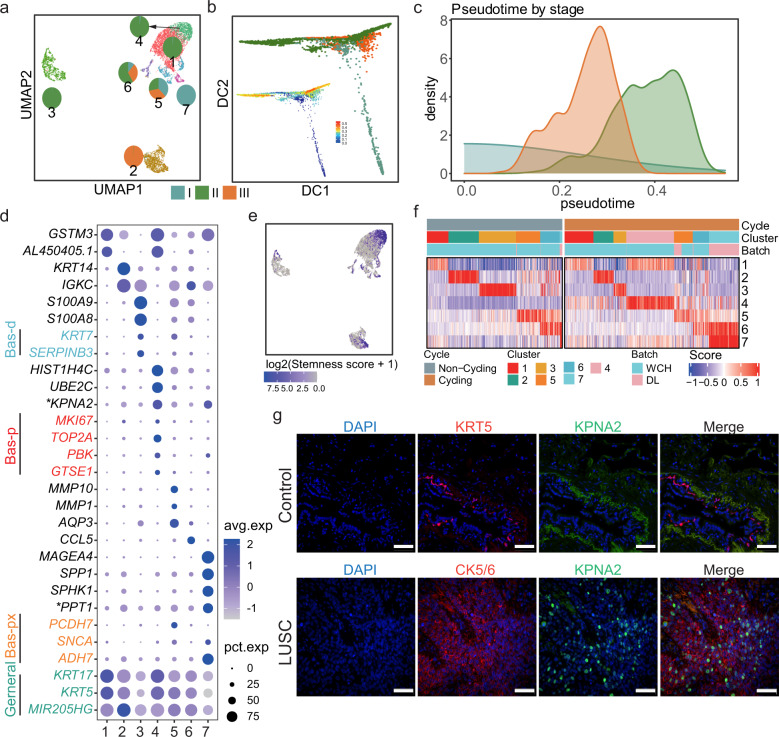
Fig. 7.
Subclusters and pseudotime analysis of the malignant basal cells in LUSC. a UMAP of 8016 malignant basal cells from LUSC, colored with different clusters, and the fraction of different stages of each cluster were labeled with a pie chart. b Diffusion map of malignant basal cells from LUSC, colored with different stages. The diffusion map colored with pseudotime was plotted at the left top corner. c Density of pseudotime colored with different stages. d Dot plot of the mean level of expression (dot intensity, blue scale) of cluster makers and indicated basal cell markers and percent of cells in population with detected expression (dot size). Generic markers colored in green, proliferation basal (Bas-p) markers colored in red, proximal basal (Bas-px) markers colored in yellow and differentiating basal (Bas-d) markers colored in blue. e UMAP plot showed the log2 transformed stemness score calculated using the mean level of expression of Bas-p markers. f Heatmap showed the cluster scores of all non-cycling cells (left) and cycling cells (right). Within each group, the cells were defined by maximal score, for cells mapping to one cluster. g The expression of KPNA2 in LUSC. Immunofluorescence staining indicated the location of KPNA2 level in lung cancer cells (green), CK5/6 (Cytokeratin 5/6) was used as the marker of lung squamous carcinoma cells (red), KRT5 (Keratin 5) was used as the marker of basal cells in normal lung, the cell nucleus was co-stained with DAPI (blue); Scale bar, 50 μm