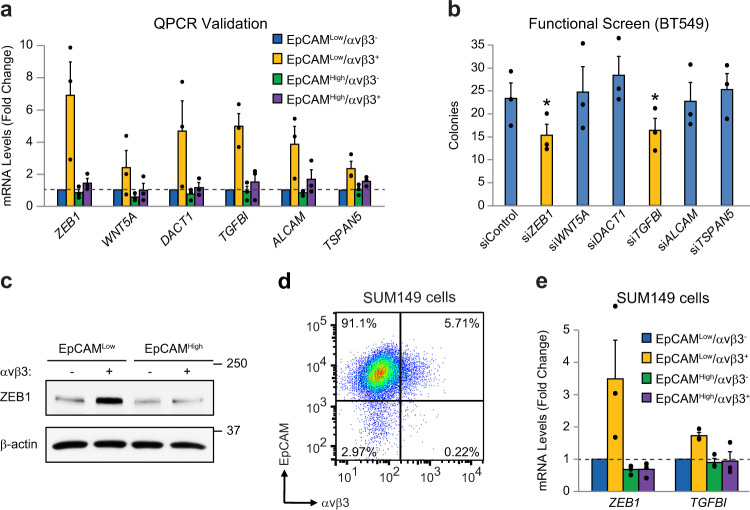
Fig. 4. Identification of TGFBI and ZEB1 as candidate genes unique to αvβ3+ CSCs.
a QPCR validation of candidate genes in sorted HCC38 cell types. b Functional screen for candidate genes necessary for methylcellulose colony formation after transient siRNA knockdown in BT549 cells. Target gene knockdown was validated by QPCR. Statistics by one-way ANOVA with Dunnett’s test. *P < 0.05. c Representative immunoblot of lysates from sorted HCC38 cells. β-actin is shown as a loading control. Molecular weight markers are indicated in kilodaltons. d Representative FACS density plot of the live, CD49f+ SUM149 cells according to their cell surface EpCAM and αvβ3 status. e QPCR validation of candidate genes in sorted SUM149 cells. a, e Samples were run in duplicate with GAPDH as a loading control. Expression is shown relative to the EpCAMLow/αvβ3− cells (dashed lines). a, b, e Data represent the mean ± s.e.m. a–e n = 3 independent experiments. See also Supplementary Fig. 4.