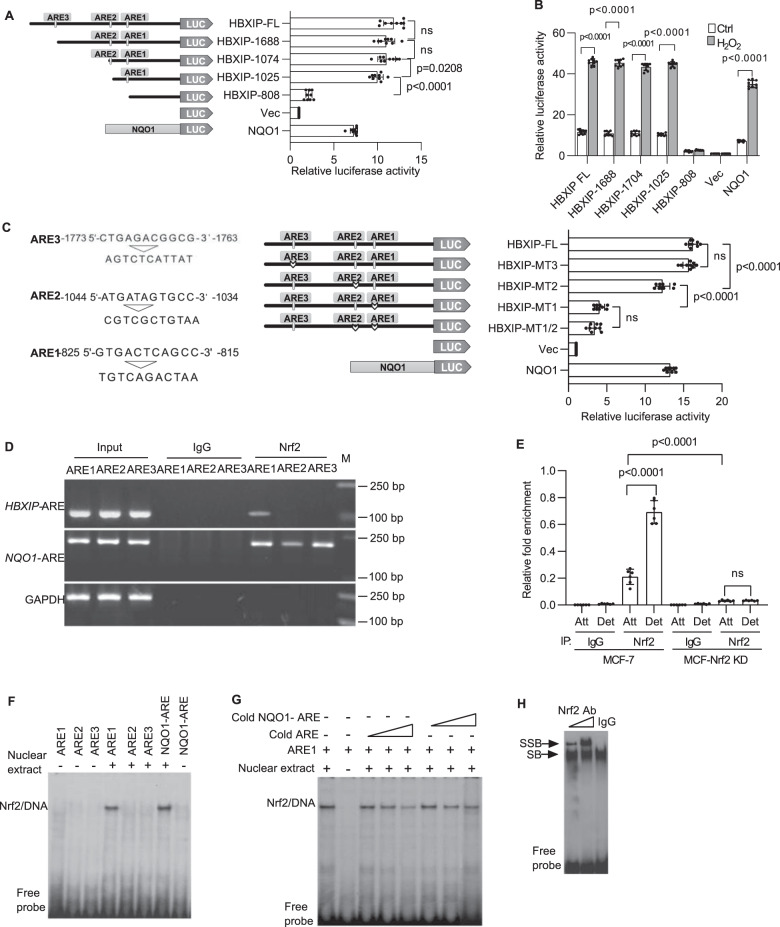
Fig. 3. Functional validation of putative Nrf2 binding sites in the HBXIP promoter.
A Systematic representation and strategy for cloning the human HBXIP gene promoter into the pGL3-Basic luciferase reporter vector. Three putative AREs (from nt -825 to -815 (ARE1), −1044 to −1034 (ARE2), and −1773 to −1763 (ARE3)) in the HBXIP promoter are shown (left panel). HBXIP promoter deletion mutants and the NQO1 promoter were separately cloned into the pGL3-Basic reporter vector and subsequently transfected into MCF-7 cells, and luciferase activity was measured (right panels). Vec, pGL3-Basic vector control. B MCF-7 cells transfected with plasmids carrying the full-length HBXIP promoter, truncated HBXIP promoter deletion mutant constructs, as indicated in A, and the NQO1 promoter was treated with DMSO or 50 μM H2O2 for 24 h, and then luciferase activity was measured. C The putative AREs were mutated as indicated (left panel). Plasmids carrying the full-length HBXIP promoter and the indicated mutated HBXIP promoters were transfected into MCF-7 cells, and luciferase activity was assessed. The human NQO1-ARE luciferase reporter plasmid was transfected into MCF-7 cells as a positive control. Vec, vector control. D ChIP assay. MCF-7 cells detached for 8 h were fixed with formaldehyde and cross-linked, and chromatin was sheared ultrasonically. Chromatin was immunoprecipitated with an anti-Nrf2 rabbit monoclonal antibody (mAb) or rabbit (DA1E) control IgG mAb. Nrf2 binding to the HBXIP promoter was analyzed by PCR with primers (Supplementary Table 2) specific for the ARE1, ARE2, and ARE3 regions in the HBXIP promoter. GAPDH primers were used as a negative control. The three ARE regions in the HBXIP promoter were amplified from 5 μl of purified soluble chromatin before immunoprecipitation for use as input DNA. The binding of Nrf2 to the NQO1-ARE promoter was used as a positive control. E ChIP and qRT-PCR. Chromatin was immunoprecipitated from attached and detached MCF-7 cells or stable MCF-Nrf2 KD cells as described in D. The binding of Nrf2 to the ARE1 region of the HBXIP promoter was measured using qRT-PCR. The amounts of immunoprecipitated DNA were normalized to those of the inputs, and the values were plotted. F HBXIP AREs (Supplementary Table 5) were end-labeled with [γ-32P]ATP and T4 kinase. Labeled DNA (100,000 cpm) was incubated with 10 μg of MCF-7 nuclear extract in binding buffer. The reaction mixtures were separated on a polyacrylamide gel and autoradiographed. DNA-protein complexes containing Nrf2 are indicated. G The nuclear proteins binding to ARE1 were competed with increasing doses of cold ARE1 or cold NQO1-ARE. H The nuclear protein/ARE1 complexes were supershifted with IgG and Nrf2 antibodies. SB shifted band, SSB supershifted band. The error bars indicate the ±SD values as assessed by Student’s t test. All experiments were performed at least three times.