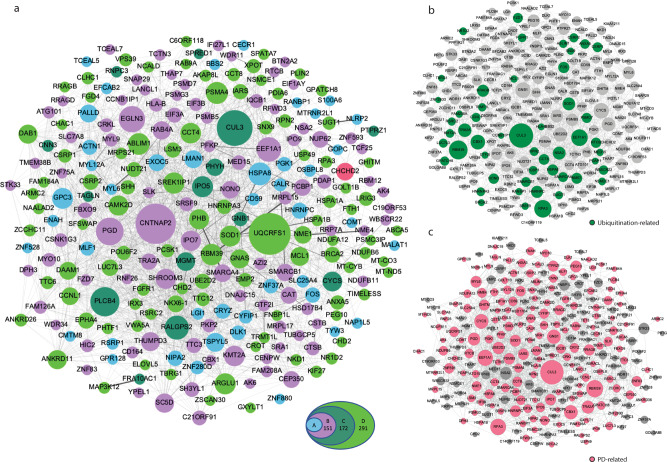
Fig. 6. Network analysis.
a Protein–protein interaction network based on known interactions available through the STRING59 and GeneMANIA61 databases. Only strong interactions were retained, predicted interactions or text associations were omitted (see Methods). Betweenness centrality was used to illustrate the relative importance of each node within the network through the level of its connectedness to other proteins. The larger the circle, the more partners the node is connected to. The colors represent the four DEG sets, with the top 56 DEGs (group A) in light blue, group B in purple, group C in dark green, and group D in light green. Each set consists of genes of the previous set plus additional genes identified by the new parameters. CHCHD2 (pink, part of group B) is a DEG, which has recently been identified as a PARK gene. Random selection of genes from genes detected by sc-RNAseq did not lead to a network formation (Supplementary Fig. 5). b DEGs which play a role in ubiquitination. Additional functional pathways are listed in Supplementary Fig. 7 and Supplementary Data 3. Specific connections to ubiquitination proteins are shown in Supplementary Fig. 8. c Based on the literature, 68% of the DEGs of this network are already known to be associated with PD (for references see Supplementary Data 4). Supplementary Fig. 9 shows which genes/proteins of the network directly interact with PARK genes through known protein–protein interactions. The topology of all three networks is the same, the different appearance is a result of a separate analysis run, but the connections and size of the nodes remain identical.