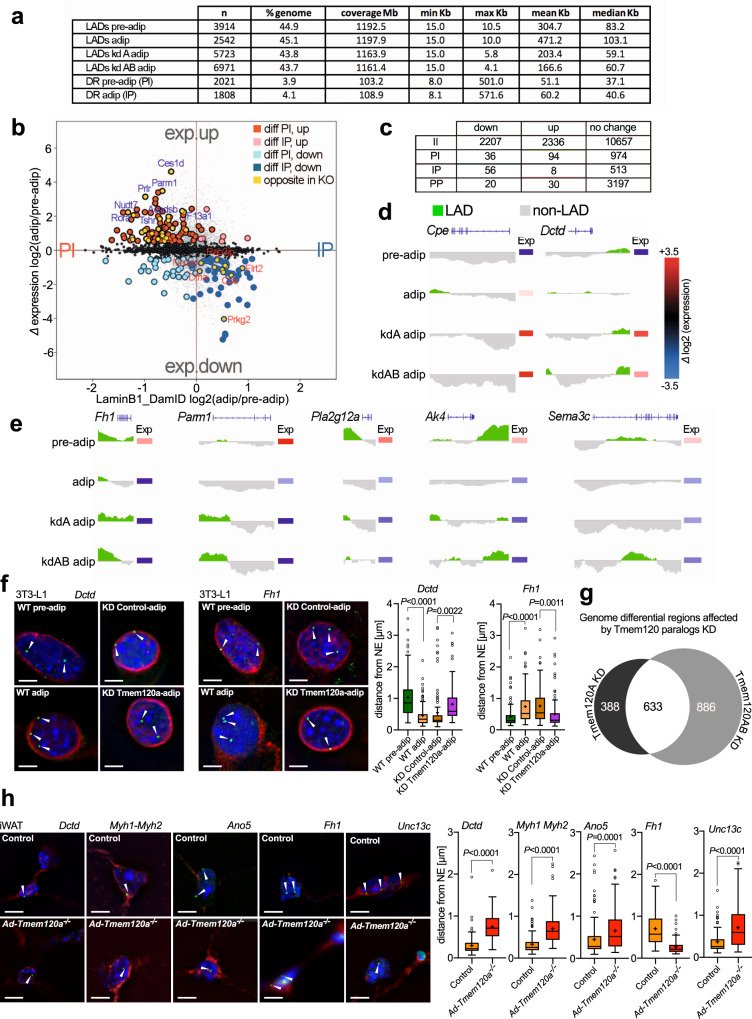
Fig. 5. DamID and transcriptomics analysis reveals Tmem120a contributes to gene regulation in adipocyte differentiation.
a LaminB1-DamID on 3T3-L1 preadipocytes and adipocytes, upon Tmem120a knockdown as well as Tmem120a and b double knockdown. Table shows metrics for lamina associated domains (LADs). b Correlation of gene expression and gene positioning shown as distribution scatter plot of genes differentially expressed (exp.up/exp.down) during adipogenesis (adip/pre-adip) and LamB1-DamID values, PI—genes moving from periphery to nuclear interion, IP—genes moving toward nuclear periphery. c Table showing numbers of genes changing expression upon being repositioned between nuclear interior “I” and nuclear periphery “P” during adipogenesis. Example DamID traces and expression changes of two genes being recruited to the NE (IP) upon differentiation that failed to reposition with Tmem120a knockdown (d), and examples for the opposite direction (PI) (e). f FISH in 3T3_L1 system for the Dctd and Fh1 genes in green with white arrow heads during wild-type adipogenesis and in the Tmem120a knockdown, distance from NE of n = 105 loci per group and gene, Tukey box plot representing median, cross on the box represents mean, bounds of box represents interquartile of the data, whiskers representing minima/maxima excluding outliers and dots represents outliers of more than 2/3 times of upper quartile, bars: 5 µm. g Venn diagram showing the number of genomic DRs affected by Tmem120 paralogs, representing the redundancy effect of single (Tmem120a) and double (Tmem120ab) knockdown on the genome organisation. h FISH in mouse iWAT from control animals vs. Ad-Tmem120a−/− detecting the position of the Dctd, Myh1/Myh2, Ano5, Fh1 and Unc13c genes; scale bars: 5 µm; distance from NE quantification is shown as Tukey box plot representing median, cross on the box represents mean, Dctd (WT n = 89 loci and KO n = 96 loci), Myh-1,2 (WT n = 105 loci, KO n = 105 loci), Ano5 (WT n = 165 loci, KO n = 138 loci), Fh1 (WT n = 111 loci, KO n = 83 loci), Unc13c (WT n = 90, KO n = 110). P values were calculated by unpaired two-sided, Student’s t test. See also Supplementary Fig. 5. Source data are provided as a Source Data file.