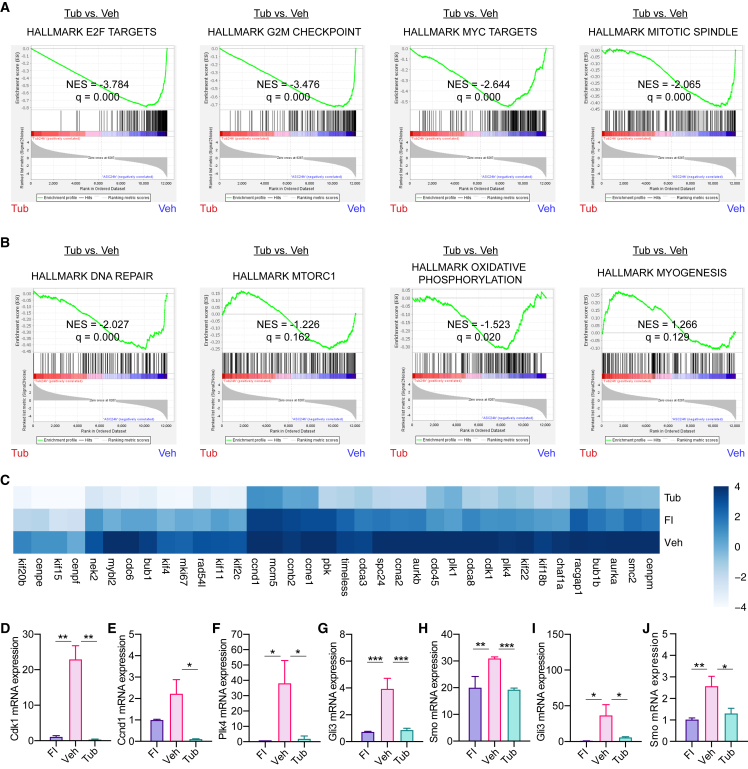
Figure 5.
Transcriptional differences between MuSCsTub and MuSCsVeh
(A and B) GSEA results for the Hallmark gene sets in comparisons of RNA-seq profiles for MuSCsTub versus MuSCsVeh after being cultured for 24 h. (A) GSEA enrichment plots for the E2F targets gene set, the G2/M checkpoint gene set, the Myc targets gene set, and the mitotic spindle gene set are shown. (B) GSEA enrichment plots for the DNA repair gene set, the mTORC gene set, the oxidative phosphorylation gene set, and the myogenesis gene set.
(C) Heatmap of differentially expressed genes between the MuSCsTub and the MuSCsVeh transcriptomes.
(D–F) RT-qPCR analyses were done in MuSCsFI, and in MuSCsVeh and MuSCsTub after being cultured for 24 h. Ct values were normalized first to the mean of Gapdh and then to the mean of MuSCsFI level in each experiment. RT-qPCR analysis of Cdk1 in (D), Ccnd1 in (E), and Plk4 in (F) (n = 3 mice).
(G and H) mRNA expression levels of Hh signaling genes such as Gli3 (G) and Smo (H) in MuSCsFI and in MuSCsVeh and MuSCsTub after being cultured for 24 h. Data obtained from our RNA-seq analysis (n = 3 mice for MuSCsFI, n = 3 mice for MuSCsVeh, and n = 4 mice for MuSCsTub).
(I and J) RT-qPCR analysis of either Gli3 (I) or Smo (J) in MuSCs. Ct values were normalized first to the mean of Gapdh and then to the mean of MuSCsFI level in each experiment (n = 6 mice). NES, normalized enrichment score in (A) and (B). Error bars represent ±s.e.m. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; no asterisk: not significant; one-tailed t test in (D)–(J).