Summary
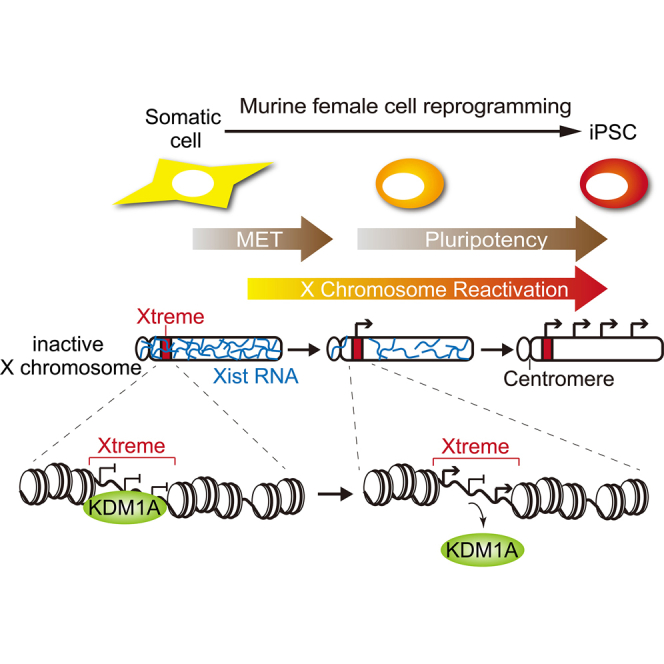
Reprogramming of murine female somatic cells to induced pluripotent stem cells (iPSCs) is accompanied by X chromosome reactivation (XCR), by which the inactive X chromosome (Xi) in female somatic cells becomes reactivated. However, how Xi initiates reactivation during reprogramming remains poorly defined. Here, we used a Sendai virus-based reprogramming system to generate partially reprogrammed iPSCs that appear to be undergoing the initial phase of XCR. Allele-specific RNA-seq of these iPSCs revealed that XCR initiates at a subset of genes clustered near the centromere region. The initial phase of XCR occurs when the cells transit through mesenchymal-epithelial transition (MET) before complete shutoff of Xist expression. Moreover, regulatory regions of these genes display dynamic changes in lysine-demethylase 1a (KDM1A) occupancy. Our results identified clustered genes on the Xi that show reactivation in the initial phase of XCR during reprogramming and suggest a possible role for histone demethylation in this process.
Keywords: X chromosome reactivation, reprogramming, epigenetics, KDM1A
Graphical abstract
Highlights
-
•
Partially reprogrammed iPSCs enabled analyses of early events in XCR
-
•
XCR initiates at a subset of genes clustered near the centromere region
-
•
XCR occurs before complete shutoff of Xist expression during reprogramming
-
•
KDM1A inhibition appears to directly reactivate transcription from the Xi
Aizawa et al. analyzed early events of X chromosome reactivation (XCR) by generation of partially reprogrammed iPSCs. They found XCR initiation at a subset of genes clustered near the centromere region before complete shutoff of Xist expression. Regulatory regions of these genes display dynamic changes in lysine-demethylase 1a (KDM1A) occupancy, suggesting a possible role for histone demethylation in this process.
Introduction
Derivation of induced pluripotent stem cells (iPSCs) from somatic cells by introduction of key pluripotency transcription factors offers tremendous potential for regenerative medicine, disease modeling, drug development, and understanding fundamental mechanisms of how cells determine their identities (Takahashi and Yamanaka, 2013). Cell identity is governed by a defined pattern of gene expression, which is underpinned by various epigenetic mechanisms (Barrero et al., 2010). As such, somatic cell reprogramming serves as an excellent model system to unravel epigenetic mechanisms that determine cell identity. One of the most drastic epigenetic changes during reprogramming is reactivation of the inactive X chromosome (XCR) in mammalian female somatic cells (Pasque and Plath, 2015). XCR is the reversal of X chromosome inactivation (XCI), which occurs during differentiation of cells in the inner cell mass (ICM) of female embryos. Because iPSCs are equivalent to embryonic stem cells (ESCs) derived from the ICM, conversion of female somatic cells into iPSCs evokes XCR when iPSCs acquire the naive pluripotent state (Nichols and Smith, 2009).
XCI and XCR involve chromosome-wide epigenetic changes during early embryonic development of female eutherian mammals. In the mouse, the paternal X chromosome undergoes imprinted XCI by the action of a non-coding RNA, Xist, at the cleavage stage (Okamoto et al., 2004) and is subsequently reactivated in the ICM of the blastocyst (Sado et al., 2001). Then ICM cells randomly choose one of the two active X chromosomes for inactivation (Lyon, 1961). The chosen X chromosome initiates expression of Xist, which subsequently spreads to coat the whole X chromosome and eventually inactivates genes on the X chromosome (Engreitz et al., 2013; Simon et al., 2013), except for escapee genes that are not subject to XCI (Brown et al., 1991). The inactive X chromosome is maintained stably by H3K27me3 and DNA methylation throughout subsequent cell divisions (Csankovszki et al., 2001).
In contrast to XCI, which is recapitulated by in vitro differentiation of female ESCs, XCR poses a major challenge for its mechanistic understanding due to lack of an appropriate in vitro system. However, XCR has been observed in several experimental systems in which somatic cells or nuclei are reprogrammed into the pluripotent state. XCR has been reported to occur in somatic cell nuclear transfer (Eggan et al., 2000), cell fusion between somatic cells and pluripotent cells (Takagi et al., 1983), and transcription factor-mediated reprogramming of somatic cells to iPSCs (Maherali et al., 2007). Studies using mouse embryos and in vitro reprogramming systems revealed a close link between XCR and pluripotency (Maherali et al., 2007). Besides the negative effect of Xist on XCR (Pasque et al., 2014), only a small number of factors are known to play a role in XCR. For example, Tsix and PRDM14 promote XCR, whereas KDM6A (also known as UTX) and HDACs delay XCR (Talon et al., 2019). A more recent study that used allele-specific RNA sequencing (RNA-seq) revealed that genes on the Xi become progressively reactivated during reprogramming (Janiszewski et al., 2019). However, the earliest events of XCR during reprogramming remain poorly defined.
We previously developed a somatic cell reprogramming system based upon replication-defective and persistent Sendai virus (SeVdp) vector (Nishimura et al., 2011). An SeVdp-derived reprogramming vector, SeVdp(fK-OSM), expresses OCT4, SOX2, KLF4, and c-MYC, and its KLF4 is tagged with a destabilization domain (DD). DD promotes degradation of the tagged KLF4, and the amount of KLF4 expressed from the vector is controllable by Shield1, a chemical inhibitor of the DD function (Nishimura et al., 2014). Depending upon the amount of Shield1 added to the culture, SeVdp(fK-OSM) converts somatic cells into iPSCs that have been reprogrammed to different extents. These partially reprogrammed iPSCs remain relatively stable and easily expandable (Nishimura et al., 2014). In this study, we used a series of partially reprogrammed iPSCs and investigated the early events of XCR during reprogramming.
Results
A TaqMan-based system to detect allele-specific transcripts from the X chromosome
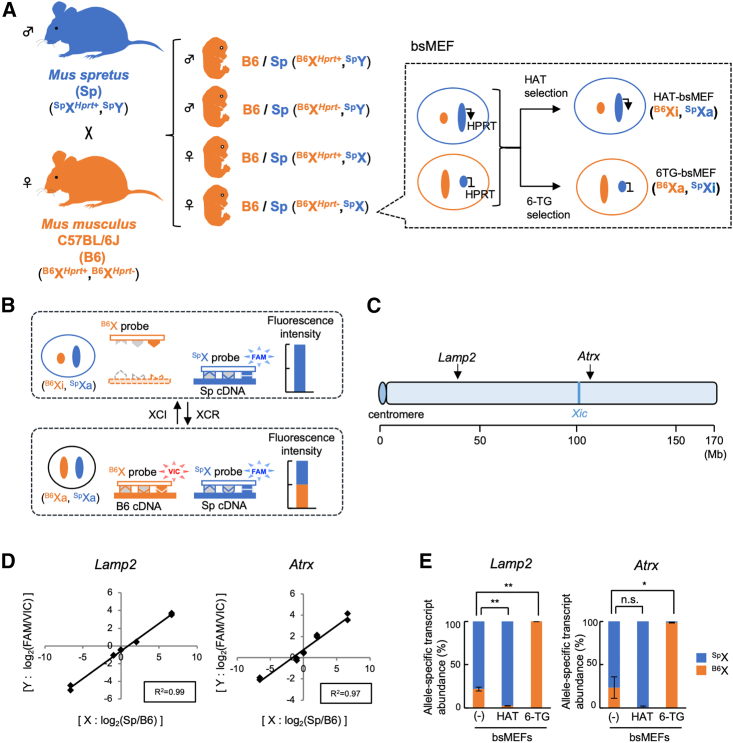
To study early events of XCR during reprogramming, we set out to establish a convenient system to monitor allele-specific transcripts from the Xi. First, we produced hybrid embryos by crossing male Mus spretus (Sp) and female Mus musculus domesticus C57BL/6J (B6) that carried a mutant Hprt allele (Figure 1A). Mus spretus and Mus musculus strains diverged more than 1.5 million years ago and thus show one sequence variant every 50–130 bases (Mahler et al., 2008; Zhang et al., 2005). Thus, hybrid cells between the two strains would possess many single-nucleotide polymorphisms (SNPs), multiple-nucleotide polymorphisms (MNPs), and indels, which are available for distinguishing allele-specific transcripts. On the basis of genome analysis of the hybrid embryos, we chose females that carried the mutant Hprt allele to obtain mouse embryonic fibroblasts (MEFs) (Figure 1A). The obtained MEFs, termed bsMEFs, were then selected with hypoxanthine-aminopterin-thymidine (HAT) or 6-thioguanine (6-TG) to isolate cells that carried a Mus musculus-derived X chromosome in an inactive (B6Xi) or active (B6Xa) form, respectively (Figure 1A). The selected cells were confirmed by allele-specific digestion patterns of the amplified Lamp2 transcripts from the X chromosome (Figures S1A and S1B).
Figure 1.
Establishment of a TaqMan-based system to quantify allele-specific transcripts from the X chromosomes
(A) Isolation of hybrid MEFs for XCR analyses.
(B) TaqMan probes that discriminate transcripts from B6 and Sp alleles. TaqMan probes in a single reaction recognize their cognate allele-specific transcripts, which differ by SNPs or an MNP, and emit fluorescence (VIC or FAM).
(C) Mouse X chromosome showing the locations of Lamp2, Atrx, and the X-inactivation center (Xic).
(D) Linear correlation between the ratio of the allele-specific cDNA amounts and the ratio of emitted fluorescence for each allele. cDNAs derived from homozygous B6 and Sp mice were mixed at five different ratios, and qRT-PCR was performed using TaqMan probes for the Lamp2 or Atrx gene. The square of the correlation coefficient is indicated by R2.
(E) Relative abundance of allele-specific transcripts in drug-selected MEFs. The relative abundances of allele-specific transcripts in HAT- or 6-TG-selected bsMEFs were calculated by using TaqMan probes for Lamp2 or Atrx.
Data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01. n.s., not significant.
We then searched a database of the Mouse Genomes Project at the Wellcome Sanger Institute (https://www.sanger.ac.uk/data/mouse-genomes-project/) for polymorphisms to design TaqMan probes that distinguish allele-specific transcripts from Sp and B6 X chromosomes (Figure 1B). Among the tested TaqMan probes, those for Lamp2 and Atrx, which included transversion in one MNP and two SNPs, respectively, were found to discriminate alleles quantitatively (Figures 1C, 1D, S1C, and S1D). These probes enabled quantitative estimation of allele-specific transcripts in highly correlative linear ranges (R2 > 0.9), which approached 2 orders of magnitude (Figures 1D and S1E). The allele-specific transcript analyses showed that unselected bsMEFs exhibited a skewed XCI, with 70%–75% of the cells possessing an active Sp X chromosome (SpXa) (Figure 1E). In addition, HAT or 6-TG selection highly enriched MEFs with B6Xi and SpXa (HAT-bsMEFs) or with B6Xa and SpXi (6TG-bsMEFs), respectively (Figures 1A and 1E). Thus, the TaqMan probes encompassing either a single MNP or two SNPs enable quantification of allele-specific transcripts from the Lamp2 and Atrx genes, providing a quick, sensitive means to capture low-level transcripts from the Xi during reprogramming.
Partially reprogrammed iPSCs permit gene expression from the Xi
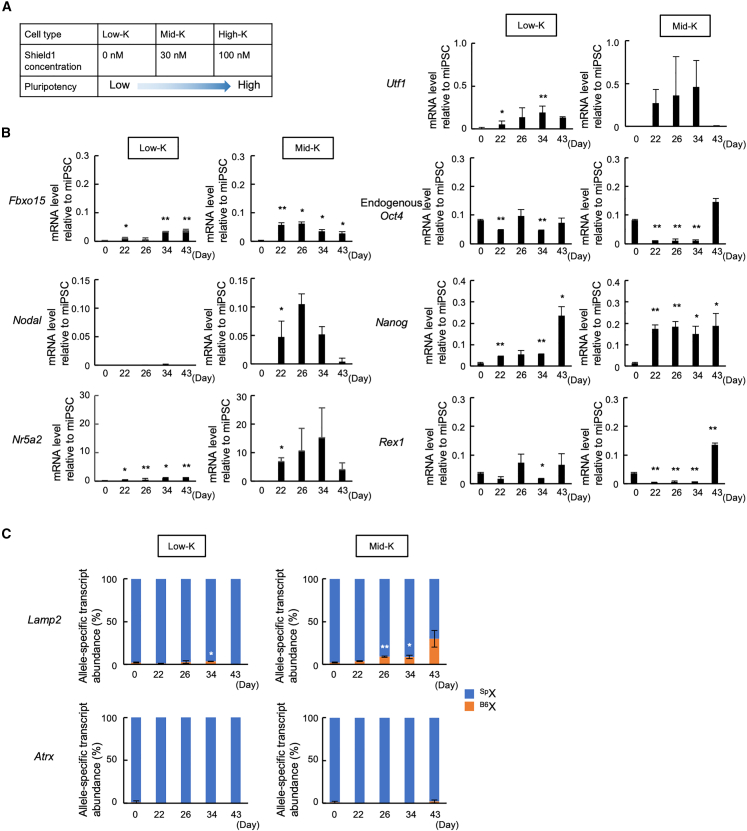
To generate iPSCs, we used SeVdp(fK-OSM) vector that expresses four reprogramming factors, OCT4, SOX2, c-MYC, and KLF4 (Nishimura et al., 2011). In this vector, KLF4 is fused with a DD, the KLF4 protein level is reduced by ubiquitin-mediated degradation (Banaszynski et al., 2006), and the KLF4 level is adjustable by the amount of added Shield1 (Nishimura et al., 2014) (Figure 2A). Indeed, addition of 100 nM Shield1 practically prevents KLF4 degradation and reprograms MEFs to the fully pluripotent state. Without Shield1, however, the KLF4 level is reduced to ∼30%, and reprogramming virtually stalls at an intermediate stage (Nishimura et al., 2014). Addition of 30 nM Shield1 increases the KLF4 level to ∼70% and allows reprogramming to a more advanced stage before stalling (Nishimura et al., 2014). Although these partially reprogrammed iPSCs are relatively stable in the SeVdp(fK-OSM) system (Nishimura et al., 2014), we noted that prolonged culture allowed them to progress in reprogramming, albeit at a markedly slower rate.
Figure 2.
Reactivation of X-linked genes from the Xi in partially reprogrammed iPSCs
(A) Generation of iPSCs with different pluripotency levels (Low-K, Mid-K, and High-K) by SeVdp(fK-OSM) in the presence of different Shield1 concentrations.
(B) Expression kinetics of pluripotency markers in partially reprogrammed iPSCs. HAT-bsMEFs were reprogrammed under the Low-K and Mid-K conditions. The expression levels of indicated genes were determined by qRT-PCR at indicated days of reprogramming.
(C) Relative abundances of allele-specific transcripts of Lamp2 and Atrx in HAT-bsMEFs reprogrammed under the Low-K and Mid-K conditions. The same cells prepared in (B) were analyzed for qRT-PCR using TaqMan probes.
All data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01 versus HAT-bsMEFs (day 0).
We reprogrammed HAT-bsMEFs by using SeVdp(fK-OSM) with 100 nM Shield1 (High-K) to generate iPSCs that express endogenous Oct4, Nanog, and Rex1 at high levels (Figure S2A). These fully reprogrammed iPSCs expressed Lamp2 and Atrx from B6Xi (Figures S2B), with clear downregulation of Xist (Figure S2C). However, the B6X-specific transcripts often surpassed the cognate SpX-specific transcripts at this stage of reprogramming (Figure S2B). This suggests that loss of an X chromosome (Pasque et al., 2018), a secondary XCI (Kim et al., 2014), or erosion of Xa (Vallot et al., 2015) may have occurred when iPSCs reached the fully pluripotent state. A similar phenomenon was also observed when we reprogrammed female MEFs derived from Momiji mice, which carries mCherry and EGFP at the Hprt locus (Kobayashi et al., 2016). When mCherry(+)/EGFP(−) MEFs were reprogrammed with SeVdp(fK-OSM) at 100 nM Shield1, 81.4% and 93.2% of the cells became mCherry(+)/EGFP(+) at days 30 and 37, respectively, which indicates that XCR occurred in these cells (Figures S2D and S2E). However, a substantial faction of cells lost expression of either mCherry or EGFP at day 44 (Figure S2E).
Given the instability of XCR when iPSCs were fully reprogrammed, we sought to focus on the earlier phase of XCR, which appeared to have initiated by day 30 of reprogramming under the High-K condition (Figure S2E). To obtain iPSCs at earlier stages, we reprogrammed MEFs without Shield1 or with 30 nM Shield1 (Figure 2A). When HAT-bsMEFs were reprogrammed by SeVdp(fK-OSM) without Shield1 (Low-K) or with 30 nM Shield1 (Mid-K) for 43 days, the cells progressed to intermediate stages and expressed various pluripotency markers, including Fbxo15, Nodal, Nr5a2, Utf1, endogenous Oct4, Nanog, and Rex1, at low or moderate levels (Figure 2B) (Nishimura et al., 2014). Low-K cells expressed low levels of Lamp2 from the Xi at day 34, and Mid-K cells expressed significant levels of Lamp2 at days 26 and 34. Both Low-K cells and Mid-K cells failed to express Atrx from the Xi until day 43 (Figure 2C). Low-K cells showed no significant Xist downregulation throughout reprogramming, whereas Mid-K cells showed Xist downregulation at day 43 (Figure S2F). These results suggest that some genes on the Xi may become reactivated at an early reprogramming stage, before acquisition of full pluripotency during reprogramming.
A subset of genes exhibits early-onset reactivation from the Xi during reprogramming
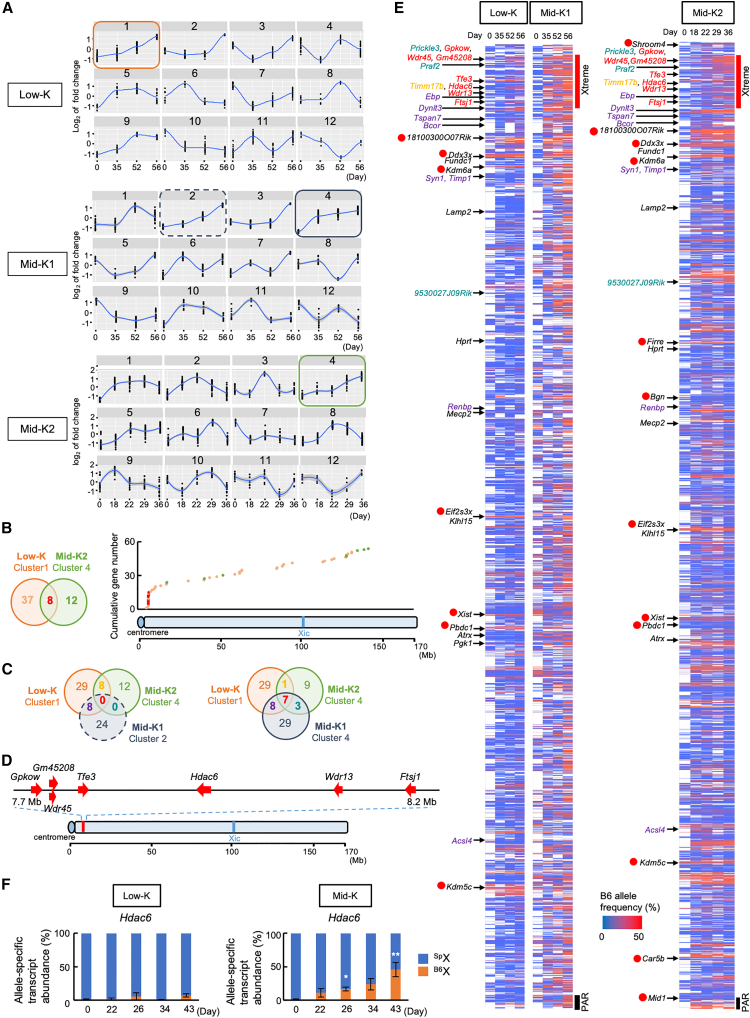
To obtain a comprehensive view of chromosome-wide gene reactivation from the Xi at intermediate stages, we performed RNA-seq analysis of Low-K and Mid-K (hereafter termed Mid-K1) cells at different time points of reprogramming (up to day 56). Besides Mid-K1 cells, we prepared Mid-K2 cells reprogrammed for a shorter period (up to day 36) because Mid-K1 cells had already expressed Lamp2 by day 35 (Figure S3A). The expression of Lamp2 expression from the Xi (Figure S3A) and pluripotency markers (Figure S3B) showed that Mid-K2 cells are intermediate between Low-K and Mid-K1 cells. After RNA-seq of Low-K, Mid-K1, and Mid-K2 cells, the X-linked transcript reads encompassing SNPs, MNPs, and indels were assigned to either B6 or Sp X chromosome. Then, the B6- or Sp-specific transcripts were annotated to more than 400 X-linked genes to quantitatively assess the relative level of B6Xi-specific transcripts (Figure S4A).
Throughout the analyzed periods of reprogramming, alleles on the Xa showed relatively constant expression levels, whereas those on the Xi showed little or low expression except for escapee genes (Figure S4B). First, we categorized gene expression patterns of B6Xi-specific transcripts into 12 distinct clusters in an unbiased manner. Low-K cluster 1, Mid-K1 clusters 2 and 4, and Mid-K2 cluster 4 included genes that gradually increased expression throughout reprogramming (Figure 3A). Eight genes in the overlap of Low-K cluster 1 and Mid-K2 cluster 4 were localized to the region proximal to the centromere (Figure 3B). Seven of them were also included in Mid-K1 cluster 4 but none in Mid-K1 cluster 2 (Figure 3C). Thus, Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1 are shared among the three clusters and localized near the centromere of the X chromosome (Figure 3D).
Figure 3.
Expression profiles of the genes on the Xi during reprogramming
(A) Clustering of expression patterns of genes reactivated on the Xi. X-linked genes were assigned into 12 clusters by expression patterns in Low-K, Mid-K1, or Mid-K2 cells. The colored squares indicate clusters that show a trajectory of increasing expression.
(B) X chromosomal locations of the genes that show increasing expression and are shared between Low-K and Mid-K2. A Venn diagram shows overlapping genes between Low-K cluster 1 and Mid-K2 cluster 4 (left panel), and a scatter diagram shows distribution of the all genes included in Low-K cluster 1 and Mid-K2 cluster 4 (right panel). The colors of dots in the scatter diagram reflect the ones in the Venn diagram.
(C) Venn diagrams show overlapping genes among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 2 (left) as well as those among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 4 (right).
(D) Locations of the seven genes that show early-onset reactivation.
(E) Heatmaps of transcriptional reactivation of the genes on the Xi chromosome. B6 allele frequencies in each polymorphic position were ordered by their genomic locations on the X chromosome. The genes that are common in among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 4 are colored in yellow (one gene), red, (seven genes), purple (eight genes), and green (three genes), as in the right diagram in (C). Genes regarded as escapee genes are marked with red circles. Xtreme region and pseudoautosomal region (PAR) are shown with red and black lines, respectively.
(F) Relative abundances of allele-specific transcripts of Hdac6 in HAT-bsMEFs reprogrammed under the Low-K and Mid-K conditions. The same cells prepared in Figure 2B were analyzed.
Data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01 versus HAT-bsMEFs (day 0).
We then calculated the relative levels of the Xi-specific transcripts against the Xa-specific transcripts at different time points of Low-K, Mid-K1, and Mid-K2 cells to generate heatmaps. As evident in Figure 3E, reactivation of genes on the Xi occurred earlier near the centromere region than in other regions. This region corresponds to the same 7.7–8.2 Mb region identified by clustering gene expression patterns (Figure 3C) and includes Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1 (Figure 3D). These genes initiated expression from the Xi when the mesenchymal markers (Cdh2, Snai1, Zeb2, and Twist1) were downregulated and epithelial markers (Cdh1, Ocln, and Epcam) were upregulated, which indicates an ongoing mesenchymal-epithelial transition (MET) (Skrypek et al., 2017) (Figure S4C). In addition, early-onset reactivation was confirmed more quantitatively by a TaqMan probe for Hdac6 (Figures 3F and S1D), which showed a similar expression profile to that by RNA-seq data (Figure 3E). By contrast, genes expressed from the Xi in HAT-bsMEFs included escapee genes (Shroom4, 1810030O07Rik, Ddx3x, Kdm6a, Firre, Bgn, Eif2s3x, Xist, Pbdc1, Kdm5c, Car5b, and Mid1), which are defined as genes that show at least 10% expression from the Xi versus Xa (Figure 3E) and reported as escapee genes by others (Brown et al., 1991; Carrel and Willard, 2005). Given that the seven genes that show early-onset reactivation were clustered in a ∼0.5 Mb region, we named the region Xtreme (X-transcriptional reactivation manifesting element).
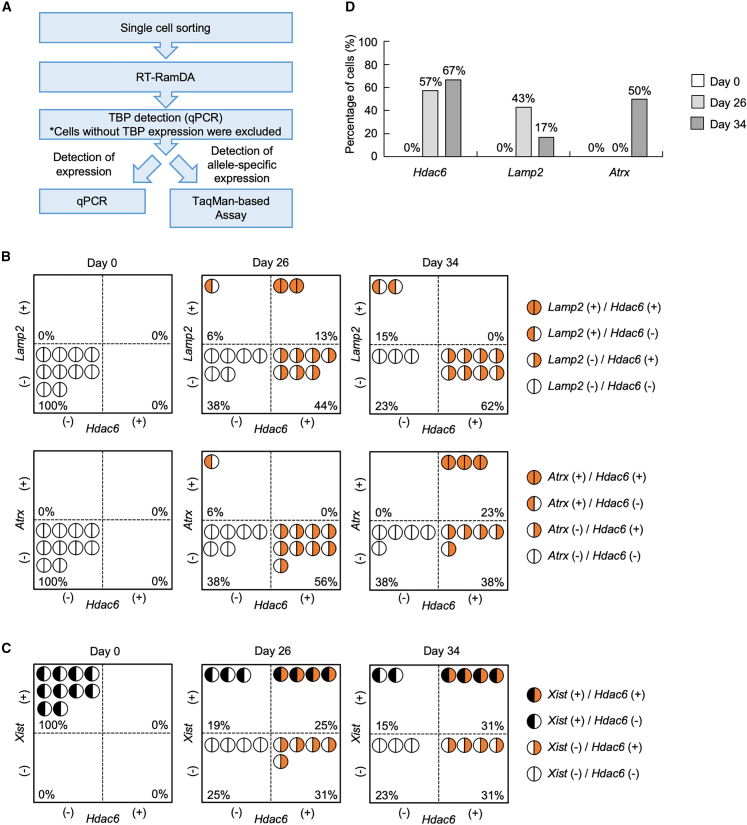
Single-cell analysis of early-onset reactivation from the Xi during reprogramming
To further confirm early-onset reactivation of genes in the Xtreme region, we used Mid-K cells to analyze Hdac6 reactivation on the Xi at single-cell resolution. MEFs were reprogrammed in the presence of 30 nM Shield1 for 26 or 34 days, and the cells were sorted using fluorescence-activated cell sorting (FACS) to obtain single cells. RNA was prepared from each cell and used to generate cDNA by reverse transcription with random displacement amplification (RT-RamDA), a method that captures transcriptome in single cells with high sensitivity (Hayashi et al., 2018). The cDNA was then used to detect allele-specific expression of Hdac6, Lamp2, and Atrx by the TaqMan-based method (Figure 4A). In addition, Xist expression in each cell was determined using qPCR of the cDNA (Figure 4A). At day 26 of reprogramming, 44% of the cells were Hdac6(+) and Lamp2(−), whereas 6% were Hdac6(−) and Lamp2(+). At day 34, 62% of the cells were Hdac6(+) and Lamp2(−), whereas 15% were Hdac6(−) and Lamp2(+) (Figure 4B, upper panels). These results show that Hdac6 has tendency to be reactivated earlier than Lamp2 from the Xi during reprogramming. Comparison of Hdac6 and Atrx also showed that Hdac6 is reactivated earlier than Atrx on the Xi (Figure 4B, lower panels). Using single-cell analysis, we also analyzed the relationship between Xist expression and Hdac6 reactivation on the Xi. At days 26 and 34 of reprogramming, 25% and 31% of the cells showed reactivation of Hdac6 while still expressing Xist (Figure 4C). However, in the cells that expressed both Hdac6 and Xist on the Xi, Xist expression was clearly reduced (Figure S5). As shown in Figure 4D, Hdac6, Lamp2, and Atrx were expressed from the Xi in 57%, 43%, and 0% of the Xist(+) cells at day 26, respectively. At day 34, Hdac6, Lamp2, and Atrx were expressed from the Xi in 67%, 17%, and 50% of the Xist(+) cells, respectively. Thus, Lamp2 and Atrx were also reactivated on the Xi in the cells expressing Xist. Collectively, these single-cell analyses confirm that Hdac6 in the Xtreme region tends to be reactivated earlier than Lamp2 and Atrx during reprogramming. In addition, a substantial fraction of cells shows reactivation of genes on the Xi even when Xist is still present.
Figure 4.
Transcriptional reactivation from the Xi and Xist expression in single cells
(A) Flowchart of single-cell analysis to detect Xist expression and allele-specific Lamp2, Atrx, and Hdac6 transcripts.
(B and C) Patterns of allele-specific gene expression in single cells. Expression of Hdac6, Lamp2, and Atrx from the Xi in single cells was determined using qRT-PCR with TaqMan probes. Xist expression was also determined from same cDNAs. Cells were categorized into indicated groups by expression patterns of Hdac6 and Lamp2 or Atrx (B) or Hdac6 and Xist (C).
(D) Cells that showed Hdac6, Lamp2, and Atrx reactivation with Xist expression. The numbers indicate percentages of cells that expressed Hdac6, Lamp2, or Atrx from the Xi in populations of Xist-expressing cells.
Raw data are shown in Figure S5.
Identification of factors required for early-onset reactivation on the Xi
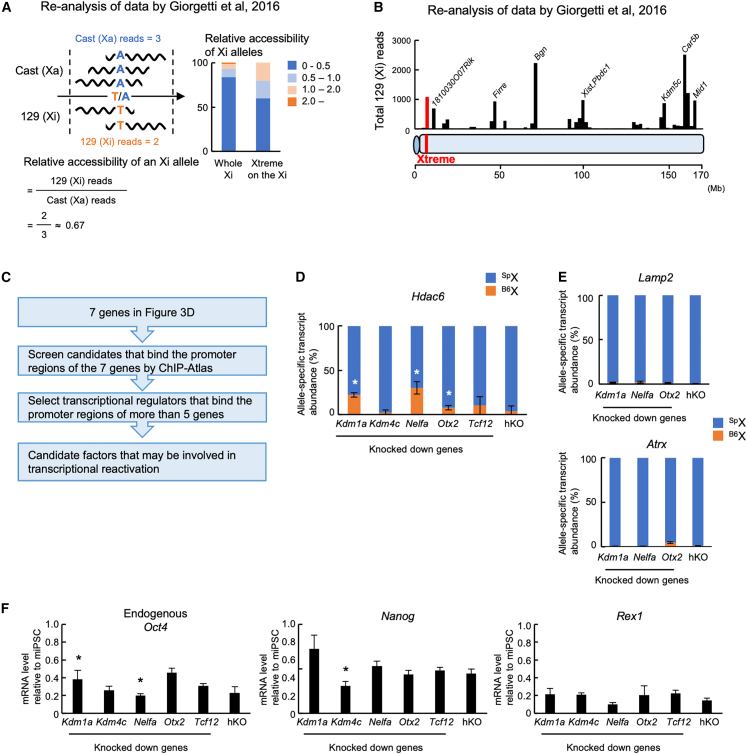
To find structural features of the Xtreme region, we re-analyzed the assay for transposase-accessible chromatin (ATAC) sequencing data of the Xi chromosome in hybrid female cells between M. m. domesticus and M. m. castaneus (Giorgetti et al., 2016). We found that the Xtreme region on the Xi adopts a more accessible structure compared with the whole Xi (Figure 5A). In addition, Figure 5B shows that the chromatin accessibility of the Xtreme region is comparable with regions containing escapees, 1810030O07Rik, Firre, Bgn, Xist, Pbdc1, Kdm5c, Car5b, and Mid1 (Yang et al., 2010). Moreover, the Xtreme region is one of the gene-rich regions on the X chromosome and forms a single TAD (topologically associating domain) in ESCs (Marks et al., 2015). These findings suggest that the Xtreme region on the Xi chromatin may be relatively accessible for transcriptional regulators during an early stage of reprogramming.
Figure 5.
Screening of transcriptional regulators potentially involved in transcriptional reactivation on the Xi
(A) Chromatin accessibility of the Xi chromosome. Left: calculation of relative accessibilities of alleles on the Xi. Right: relative accessibilities of the alleles in the Xtreme region in the Xi. The relative accessibilities of alleles were classified into four groups according to their values. Percentages of the four groups are indicated for the whole Xi or the Xtreme region of the Xi.
(B) Variation in chromatin accessibility in the whole Xi. 129 (Xi) reads that overlap with each polymorphism were summed up in 2 Mbs windows and shown along X chromosome with the Xtreme region and escapee genes.
(C) Flowchart to screen for transcriptional regulators that may be involved in transcriptional reactivation on the Xi.
(D and E) Relative abundance of allele-specific transcripts when the candidate genes were knocked down. After retrovirus-mediated transduction of shRNA against each candidate, HAT-bsMEFs were reprogrammed under the Mid-K condition. Relative abundance of allele-specific transcripts for Hdac6 (D) or Lamp2 and Atrx (E) were determined at about day 20 of reprogramming.
(F) Expression of pluripotency markers in HAT-bsMEFs reprogrammed with knockdown of the candidate genes. Endogenous Oct4, Nanog, and Rex1 mRNA levels were determined in the same cells used in (D).
All data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 versus reprogrammed cells with control hKO-expressing retrovirus infection.
To screen for such factors that might gain access to the Xtreme region and function in the reactivation of the genes during an early stage of reprogramming, we used the ChIP-Atlas database (Oki et al., 2018), which includes 332 transcriptional regulators that have been experimentally confirmed to occupy the regulatory regions within the Xtreme region in ESCs. We searched for factors that show occupancy within the 1 kb of the transcription start sites (TSSs) of Gpkow, Gm45208, Wdr45, Tfe3, Hdac6, Wdr13, and Ftsj1 genes (Figures 5C and S6A–S6C), all of which show early-onset reactivation on the Xi during reprogramming (Figure 3D). Five transcriptional regulators, KDM1A, KDM4C, NELFA, TCF12, and OTX2, were found to occupy the regulatory regions of at least five of the seven genes in the Xtreme region (Figures 5C and S6A–S6C).
To confirm their involvement in early-onset reactivation of the genes on the Xi, we knocked down each candidate gene by short hairpin RNAs (shRNAs) expressed from silencing-resistant retrovirus vectors (Figures S6D–S6G) and reprogrammed the cells with SeVdp(fK-OSM) with 30 nM Shield1. After 20 days of reprogramming, almost no reactivation of Hdac6 on the Xi was observed in the cells transduced with a control retrovirus expressing a fluorescent protein, Kusabira-Orange (hKO). However, knockdown of Kdm1a, Nelfa, or Otx2 elicited Hdac6 reactivation to various but statistically significant extents (Figure 5D). By contrast, Lamp2 and Atrx were not reactivated on the Xi at this stage of reprogramming (Figure 5E). Importantly, expression of shRNA for Kdm1a, Nelfa, or Otx2 showed only minor effects on the expression levels of pluripotency markers such as endogenous Oct4, Nanog, and Rex1 (Figure 5F), indicating that knockdown of Kdm1a, Nelfa, or Otx2 may facilitate Hdac6 reactivation even when they do not promote the reprogramming process. These results suggest a possible role for KDM1A, NELFA and OTX2 in facilitating early-onset reactivation of genes on the Xi.
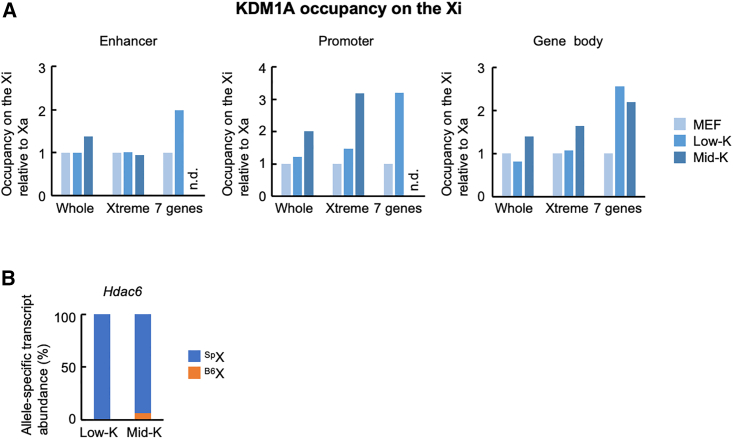
Dynamic changes of KDM1A occupancy upon reactivation of genes in the Xtreme region of the Xi
Given that the seven genes in the Xtreme region retain relatively accessible chromatin on the Xi (Marks et al., 2015) (Figures 5A and 5B), we hypothesized that these genes may be reactivated via conformational and/or epigenetic regulation of chromatin. We therefore focused on an epigenetic regulator, KDM1A (also known as LSD1), and performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis of HAT-bsMEFs with or without reprogramming under the Low-K or Mid-K conditions. We then determined KDM1A occupancies on the Xi relative to Xa and compared KDM1A occupancies on the seven genes, the Xtreme region, and the whole Xi. Under the Low-K condition, KDM1A occupancies on the enhancer, promoter, and gene body of the seven genes increased noticeably compared with the Xtreme region or the whole Xi chromosome (Figure 6A). Under the Mid-K condition, KDM1A occupancy virtually disappeared at the enhancer and promoter regions, concomitant with partial reactivation of Hdac6 (Figure 6B). Even under this condition, however, KDM1A occupancy at the gene body remained relatively high. Thus, these dynamic changes in KDM1A occupancy suggest a possible physical and/or functional regulation of KDM1A during reactivation in the Xtreme region of the Xi.
Figure 6.
KDM1A occupancy in the Xtreme region
(A) KDM1A occupancy in different XCR states. HAT-bsMEFs were reprogrammed under the Low-K or Mid-K condition for about 20 days. The extracted chromatins were used for allele-specific ChIP-seq for KDM1A. KDM1A occupancies on the Xi relative to its occupancies on the Xa at enhancer, promoter, and gene body are shown. Relative KDM1A occupancies on the Xi in MEFs are set to 1. Seven genes are Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1 in the Xtreme region. n.d., not detected.
(B) Relative abundance of allele-specific Hdac6 transcripts in the cells used for ChIP-seq.
Data consist of a single experiment.
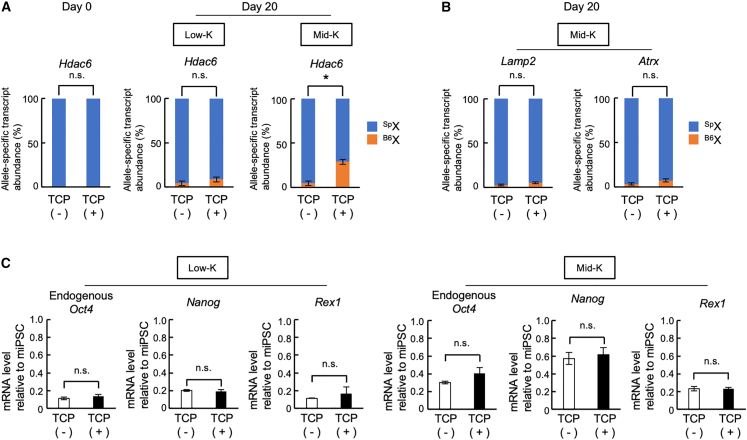
To further confirm the role of KDM1A in the reactivation, as indicated by shRNA experiments (Figure 5D), we tested tranylcypromine (TCP), a specific inhibitor of the histone demethylase activity of KDM1A (Sun et al., 2016). HAT-bsMEFs were reprogrammed for 20 days with SeVdp(fK-OSM) in the presence or absence of 5 μM TCP under the Low-K or Mid-K condition. When HAT-bsMEFs were reprogrammed under the Low-K condition, TCP did not enhance expression of Hdac6 from the Xi (Figure 7A). Under the Mid-K condition, however, TCP significantly enhanced expression of Hdac6 from the Xi (Figure 7A) but not Lamp2 and Atrx2 (Figure 7B), suggesting that inhibition of the histone demethylase activity facilitates reactivation of Hdac6 on the Xi. Although TCP promotes reprogramming when retrovirus is used for expressing reprogramming factors (Sun et al., 2016), TCP did not promote Sendai virus-mediated reprogramming under the Low-K or Mid-K condition (Figure 7C). Thus, KDM1A inhibition appears to directly reactivate Hdac6 expression from the Xi rather than via indirect effect through promotion of reprogramming.
Figure 7.
Enhanced transcriptional reactivation on the Xi by a KDM1A inhibitor, TCP
(A and B) Effect of TCP on the transcriptional reactivation of the Xi. HAT-bsMEFs were reprogrammed under the Low-K or Mid-K condition with or without 5 μM TCP. Abundance of allele-specific Hdac6 transcripts at days 0 and 20 of reprogramming (A) and Lamp2 and Atrx transcripts at day 20 of reprogramming (B) were determined using qRT-PCR with TaqMan probes.
(C) Effect of TCP on induction of pluripotency markers. Endogenous Oct4, Nanog, and Rex1 mRNA levels were determined at day 20 of reprogramming as described in (A).
All data represent mean ± SEM of at least three independent experiments. ∗p < 0.05. n.s., not significant.
Discussion
XCR is observed during female cell reprogramming via introduction of transcription factors into somatic cells (Maherali et al., 2007; Tran et al., 2018) and fusion of somatic cells with ESCs (Tada et al., 2001). These studies underscored a close coupling between XCR and pluripotency, and this coupling is believed to be mediated by the pluripotency factors OCT4, SOX2, and NANOG, which bind the first intron of Xist to downregulate its expression (Navarro et al., 2010). However, these studies assessed XCR by relying on fluorescence in situ hybridization (FISH) analysis of the Xist RNA (Chaumeil et al., 2003) or reactivation of well-studied X-linked genes such as Pgk1, Mecp2, and Atrx (Maherali et al., 2007; Pasque et al., 2014; Tomoda et al., 2012). In our RNA-seq analyses of partially reprogrammed iPSCs, Pgk1, Mecp2, and Atrx are reactivated relatively late during the process of XCR (Figure 3E), suggesting that XCR in previous studies generally designates its completion rather than its initiation.
Although numerous reports indicate a close association between Xist shutoff and XCR, it is well known that Xist does not play an essential role in maintenance of the Xi (Brown and Willard, 1994; Csankovszki et al., 1999). Moreover, Xist is downregulated gradually after acquisition of pluripotency (Do et al., 2008; Kim et al., 2015). Indeed, some genes initiate reactivation on the Xi in the blastocyst even in the presence of Xist coating and H3K27me3 (Williams et al., 2011). In nascent primordial germ cells, some X-linked genes initiate biallelic expression before Xist expression is shut off, and XCR appears to occur over a prolonged period (Sugimoto and Abe, 2007). During generation of human iPSCs, the Xi undergoes structural changes in the presence of Xist coating (Tchieu et al., 2010). Consistent with these observations, our single-cell analysis suggests that complete shutoff of Xist is not necessary for initiating gene reactivation on the Xi. However, given that the Xist level is reduced to some extent in the cells that showed reactivation of Hdac6, Lamp2, or Atrx, it may be possible that Xist is partially removed from the Xi regions where reactivation occurs.
Our analysis of gene expression patterns indicates that some genes in the Xtreme region of the Xi initiate reactivation early when the cells are undergoing MET. This result is in good agreement with a more recent study involving allele-specific RNA sequence analysis of female hybrid cells formed between M. m. domesticus and M. m. castaneus during reprogramming (Janiszewski et al., 2019). In addition to the findings of Janiszewski et al., we could identify a cluster of genes with early reactivation on the Xi. This may be because we used a sensitive RT-PCR-based TaqMan system to select a series of stable partially reprogrammed iPSCs generated from hybrid cells between M. m. domesticus and M. spretus. However, our results are based upon partially reprogrammed iPSCs generated by an SeVdp-based vector and therefore warrant further analyses using iPSCs generated by a more commonly used retroviral system.
The Xtreme region is located near the centromere embedded in repressive heterochromatin structure (Kerry, 2014). On the mouse X chromosome, the Xtreme region is farthest from the Xist gene, from which Xist spreads in two waves to coat the whole X chromosome in cis (Simon et al., 2013). More relevantly, the Xtreme region is one of the regions where the gene density is highest on the whole X chromosome (Marks et al., 2015). Thus, the Xtreme region may form a less compact heterochromatin on the Xi. In fact, when XCI is induced by inhibiting Tsix transcription during differentiation of ESCs into NPCs, the genes in the Xtreme region of the Xi escape XCI (Marks et al., 2015), although these genes are not considered as canonical escapee genes (Berletch et al., 2015; Carrel and Willard, 2005; Li et al., 2012). Heterochromatin structure on the Xi requires repeated maintenance by regenerating heterochromatin after each DNA replication (Allshire and Madhani, 2018). If the Xtreme region is intrinsically less compact, it may become more susceptible to reactivation of the resident genes because of accelerated DNA replication and cell division at the MET stage of reprogramming (Plath and Lowry, 2011).
Although Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1 in the Xtreme region show early XCR, we note that not all genes in this region are reactivated early. Thus, in addition to regulation by changes in gross chromatin structure, each gene should be regulated individually, probably via epigenetic regulation such as histone modifications. Indeed, active removal of H3K27me3 and deposition of H3K4me3 correlate with reversal of imprinted XCI (Borensztein et al., 2017), and HDAC1/3 inhibitor enhances XCR, likely via enhanced acetylation of histones (Janiszewski et al., 2019). Our demonstration of dynamic changes in KDM1A occupancy also points to a role for histone modifications in early-onset reactivation on the Xi. KDM1A apparently substitutes some inhibitory mechanism, such as a repressive chromatin structure, and takes over a predominant role in maintaining gene repression. If this shift in repressive mechanism occurs, it would render the Xtreme region more dependent on KDM1A than other regions of the Xi. Subsequent removal of KDM1A from the regulatory regions correlates with reactivation of Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1, where KDM1A occupancy is frequently observed in ESCs (Figure S6C). Consistent with this possible role of KDM1A, the Xi was reported to lack H3K4 methylations, including H3K4me1 and H3K4me2, which are demethylated by KDM1A (Heard et al., 2001; Rens et al., 2010). Thus, restoration of H3K4me2 by either removal or inhibition of KDM1A is expected to play an important role in reactivating genes during XCR.
Experimental procedures
XCR induction by reprogramming
HAT-bsMEFs were reprogrammed by infection with SeVdp(fK-OSM) including blasticidin S-resistant gene at 32°C for 14 h, as described previously (Nishimura et al., 2014). To minimize contamination of feeder cells, feeder cells were removed by addition of 1 μg/mL of blasticidin S (Wako) 2 days before harvesting the cells. For knockdown of a gene, HAT-bsMEFs were transduced with a retroviral vector expressing shRNA against each target gene for 2 days, followed by selection of the infected cells by 1 μg/mL puromycin. To inhibit the enzymatic activity of KDM1A, HAT-bsMEFs were cultured in the presence of 5 μM of TCP hydrochloride (Abcam) from 7 days before reprogramming.
Determination of allele-specific transcripts by qRT-PCR using TaqMan probes
To quantify relative amount of allele-specific transcripts, standard cDNAs and mixtures of B6 cDNA and Sp cDNA in various ratios were prepared by mixing each cDNA at B6/Sp = 1:99, 20:80, 50:50, 66:33, or 99:1 by copy number. For the standard curve, log2(Sp cDNA copy number/B6 cDNA copy number) and log2(FAM ΔRn/VIC ΔRn) were plotted on the x and y axes, respectively (Lo et al., 2003). qPCR was performed using TaqMan Genotyping Master Mix (Thermo Fisher Scientific) using the “Genotyping” method in QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific), following a modified program: the pre- and post-PCR read was done at 25°C. The cycle number was increased to 60 cycles when the target was Hdac6. The sequences of the primers and probes are listed in Table S1.
Determination of allele-specific transcripts using RNA-seq
Variant calling was performed using the Basic Variant Call tool in CLC Genomics Workbench version 10.1.1 (Qiagen) to obtain a list of SNP positions and allele frequencies of variants in the RNA-seq reads. The B6 reference read was defined as a read aligned to the reference genome (mm10). The Sp variant read was defined as a read not assigned to the reference genome. To distinguish sequence errors from true polymorphisms, only variants found in more than two samples were classified as polymorphisms between B6 and Sp. At each polymorphism, B6 and Sp allele frequencies were calculated as B6 reference read number/(B6 reference read number + Sp variant read number) and Sp variant read number/(B6 reference read number + Sp variant read number), respectively. B6 gene frequency was calculated as an average of B6 allele frequencies of all polymorphisms in the gene. For clustering according to the B6 gene frequency profile, we used Ward’s agglomerative hierarchical clustering on the basis of a Euclidean distance matrix computed using the standard R functions hclust() with method = “ward.D2” and dist() with method = “euclidean.” Data at each time point were derived from a single sample.
Single-cell analysis of allele-specific expression
Cells were dissociated with 0.5 g/L trypsin-EDTA (Nacalai Tesque) and suspended in culture medium. Dissociated cells were subjected to single-cell sorting using MoFlo XDP (Beckman) followed by cDNA synthesis using the RT-RamDA cDNA Synthesis Kit (Toyobo). RT-RamDA cDNAs were prepared from 32 and 24 single cells of MEFs and reprogrammed cells, respectively, and only RT-RamDA cDNAs in which Tbp expression was detected using qPCR were used for further experiments. For Tbp and Xist RNA detection, one-fifth the amount of cDNA was used for qPCR analyses. For allele-specific detection using the TaqMan probe, one-fifth the amount of RT-RamDA cDNA was pre-amplified by PCR using AmpliTaq Gold 360 Mastermix with the primers listed in Table S1. PCR cycles for pre-amplification were as follows: 16 cycles for Lamp2, 20 cycles for Atrx, and 30 cycles for Hdac6. Then, one-fifth the amount of pre-amplified PCR products were used for qPCR with the TaqMan probe. Cells that exhibited more than 25% of the B6 allele-specific transcript abundance were regarded to show transcriptional reactivation of the gene on the Xi.
Statistical analysis
Student’s t tests were used to test for statistical significance difference between datasets. A p value < 0.05 was considered to indicate statistical significance.
Author contributions
S.A., K.N., and K.H. designed the research. S.A., K.N., P.L.B., Y.T.H.T., M.M., T.N., A.S., E.S., and T.S. collected and analyzed the data. S.A., K.N., G.S.M., A. Kuno, M.M., and Y.H. performed the bioinformatic analyses. S.A., A. Kumar, and S.K. prepared the materials. S.A., K.N., A.F., and K.H. wrote the paper.
Conflict of interests
The authors declare no competing interests.
Acknowledgments
Hprt-KO Mus musculus (RBRC02467) and Mus spretus (RBRC00208) were provided by the RIKEN BioResource Research Center (BRC), which is participating in National Bio-Resources Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. We thank Dr. M. Katsuki for producing the Hprt-KO Mus musculus strain. This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI grants JP19J11416 (S.A), JP16K07244 (K.N.), JP19H03203 (K.N.), JP19K22945 (K.N.), JP19K07343 (A.F.), JP17H05063 (Y.H.), JP17H04036 (K.H.), and JP21H02678 (K.H.), and the Takeda Science Foundation (K.N.). We are grateful to Y. Yamazaki and T. Nishimura for technical assistance with cell sorting and data collection, respectively.
Published: December 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.11.008.
Contributor Information
Ken Nishimura, Email: ken-nishimura@md.tsukuba.ac.jp.
Koji Hisatake, Email: kojihisa@md.tsukuba.ac.jp.
Supplemental information
Data and code availability
The accession number for the RNA-seq and ChIP-seq datasets reported in this paper is GSE157484.
References
- Allshire R.C., Madhani H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Chen L., Maynard-Smith L.A., Ooi A.G.L., Wandless T.J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero M.J., Boué S., Izpisúa Belmonte J.C. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell. 2010;7:565–570. doi: 10.1016/j.stem.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Berletch J.B., Ma W., Yang F., Shendure J., Noble W.S., Disteche C.M., Deng X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11:1–26. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M., Okamoto I., Syx L., Guilbaud G., Picard C., Ancelin K., Galupa R., Diabangouaya P., Servant N., Barillot E., et al. Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass. Nat. Commun. 2017;8:1297–1310. doi: 10.1038/s41467-017-01415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., Willard H.F. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Chaumeil J., Okamoto I., Heard E. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 2003;376:405–419. doi: 10.1016/S0076-6879(03)76027-3. [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Panning B., Bates B., Pehrson J.R., Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A location but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Nagy A., Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 2001;153:773–783. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J.T., Han D.W., Gentile L., Sobek-Klocke I., Stehling M., Schöler H.R. Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells. 2008;26:2821–2831. doi: 10.1634/stemcells.2008-0482. [DOI] [PubMed] [Google Scholar]
- Eggan K., Akutsu H., Hochedlinger K., Rideout W., Yanagimachi R., Jaenisch R. X-Chromosome inactivation in cloned mouse embryos. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S., et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1–9. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L., Lajoie B.R., Carter A.C., Attia M., Zhan Y., Xu J., Chen C.J., Kaplan N., Chang H.Y., Heard E., et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Ozaki H., Sasagawa Y., Umeda M., Danno H., Nikaido I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 2018;9:619. doi: 10.1038/s41467-018-02866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Rougeulle C., Arnaud D., Avner P., Allis C.D., Spector D.L. Methylation of histone H3 at Lys-9 Is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Janiszewski A., Talon I., Chappell J., Collombet S., Song J., De Geest N., To S.K., Bervoets G., Marin-bejar O., Provenzano C., et al. Dynamic reversal of random X-Chromosome inactivation during iPSC reprogramming. Genome Res. 2019;29:1659–1672. doi: 10.1101/gr.249706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry S.B. Centromeric heterochromatin: the primordial segregation machine. Annu. Rev. Genet. 2014;48:457–484. doi: 10.1146/annurev-genet-120213-092033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Choi H.W., Araúzo-Bravo M.J., Schöler H.R., Do J.T. Reactivation of the inactive X chromosome and posttranscriptional reprogramming of Xist in iPSCs. J. Cell Sci. 2015;128:81–87. doi: 10.1242/jcs.154294. [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Hysolli E., Tanaka Y., Wang B., Jung Y.W., Pan X., Weissman S.M., Park I.H. X chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Rep. 2014;2:896–909. doi: 10.1016/j.stemcr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Hosoi Y., Shiura H., Yamagata K., Takahashi S., Fujihara Y., Kohda T., Okabe M., Ishino F. Live imaging of X chromosome reactivation dynamics in early mouse development can discriminate naïve from primed pluripotent stem cells. Development. 2016;143:2958–2964. doi: 10.1242/dev.136739. [DOI] [PubMed] [Google Scholar]
- Li S.M., Valo Z., Wang J., Gao H., Bowers C.W., Singer-Sam J. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mahler K.L., Fleming J.L., Dworkin A.M., Gladman N., Cho H.Y., Mao J.H., Balmain A., Toland A.E. Sequence divergence of Mus spretus and Mus musculus across a skin cancer susceptibility locus. BMC Genomics. 2008;9:1–12. doi: 10.1186/1471-2164-9-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kerstens H.H.D., Barakat T.S., Splinter E., Dirks R.A.M., van Mierlo G., Joshi O., Wang S.Y., Babak T., Albers C.A., et al. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16:1–20. doi: 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Oldfield A., Legoupi J., Festuccia N., Dubois A.S., Attia M., Schoorlemmer J., Rougeulle C., Chambers I., Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Sano M., Ohtaka M., Furuta B., Umemura Y., Nakajima Y., Ikehara Y., Kobayashi T., Segawa H., Takayasu S., et al. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Kato T., Chen C., Oinam L., Shiomitsu E., Ayakawa D., Ohtaka M., Fukuda A., Nakanishi M., Hisatake K. Manipulation of KLF4 expression generates iPSCs paused at successive stages of reprogramming. Stem Cell Rep. 2014;3:915–929. doi: 10.1016/j.stemcr.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C. ChIP -Atlas: a data-mining suite powered by full integration of public ChIP -seq data. EMBO Rep. 2018;19:1–10. doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Plath K. X chromosome reactivation in reprogramming and in development. Curr. Opin. Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Karnik R., Chronis C., Petrella P., Langerman J., Bonora G., Song J., Vanheer L., Dimashkie A.S., Meissner A., et al. X chromosome dosage influences DNA methylation dynamics during reprogramming to mouse iPSCs. Stem Cell Rep. 2018;10:1537–1550. doi: 10.1016/j.stemcr.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Tchieu J., Karnik R., Uyeda M., Sadhu Dimashkie A., Case D., Papp B., Bonora G., Patel S., Ho R., et al. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K., Lowry W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W., Wallduck M.S., Lovell F.L., Ferguson-Smith M.A., Ferguson-Smith A.C. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc. Natl. Acad. Sci. U S A. 2010;107:17657–17662. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T., Wang Z., Sasaki H., Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Simon M.D., Pinter S.F., Fang R., Sarma K., Rutenberg-Schoenberg M., Bowman S.K., Kesner B.A., Maier V.K., Kingston R.E., Lee J.T. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G. Epithelial-to-Mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33:943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:1309–1317. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Liang L., Li Y., Feng C., Li L., Zhang Y., He S., Pei D., Guo Y., Zheng H. Lysine-specific histone demethylase 1 inhibition promotes reprogramming by facilitating the expression of exogenous transcriptional factors and metabolic switch. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep30903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takagi N., Yoshida M.A., Sugawara O., Sasaki M. Reversal of X-inactivation in female mouse somatic cells hybridized with murine teratocarcinoma stem cells in vitro. Cell. 1983;34:1053–1062. doi: 10.1016/0092-8674(83)90563-9. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induced pluripotent stem cells in medicine and biology. Development. 2013;140:2457–2461. doi: 10.1242/dev.092551. [DOI] [PubMed] [Google Scholar]
- Talon I., Janiszewski A., Chappell J., Vanheer L., Pasque V. Recent advances in understanding the reversal of gene silencing during X chromosome reactivation. Front. Cell Dev. Biol. 2019;7:1–13. doi: 10.3389/fcell.2019.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sean P., Aimiuwu O., Lindgren A., Hakimian S., Zack J.A., et al. Female human iPS cells retain inactive X-chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K., Takahashi K., Leung K., Okada A., Narita M., Yamada N.A., Eilertson K.E., Tsang P., Baba S., White M.P., et al. Derivation conditions impact x-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.H.Y., Fukuda A., Aizawa S., Bui P.L., Hayashi Y., Nishimura K., Hisatake K. Live cell imaging of X chromosome reactivation during somatic cell reprogramming. Biochem. Biophys. Rep. 2018;15:86–92. doi: 10.1016/j.bbrep.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C., Ouimette J.F., Makhlouf M., Féraud O., Pontis J., Côme J., Martinat C., Bennaceur-Griscelli A., Lalande M., Rougeulle C. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Williams L.H., Kalantry S., Starmer J., Magnuson T. Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development. 2011;138:2049–2057. doi: 10.1242/dev.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Babak T., Shendure J., Disteche C.M. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wheeler D.A., Yakub I., Wei S., Sood R., Rowe W., Liu P.P., Gibbs R.A., Bsjetow K.H. SNPdetector: a software tool for sensitive accurate SNP detection. PLoS Comput. Biol. 2005;1:395–404. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq and ChIP-seq datasets reported in this paper is GSE157484.