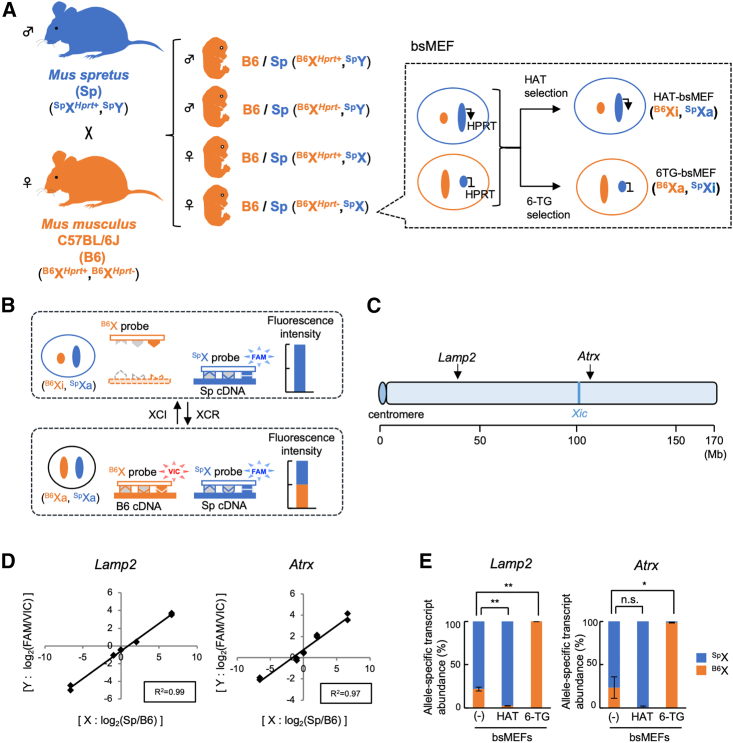
Figure 1.
Establishment of a TaqMan-based system to quantify allele-specific transcripts from the X chromosomes
(A) Isolation of hybrid MEFs for XCR analyses.
(B) TaqMan probes that discriminate transcripts from B6 and Sp alleles. TaqMan probes in a single reaction recognize their cognate allele-specific transcripts, which differ by SNPs or an MNP, and emit fluorescence (VIC or FAM).
(C) Mouse X chromosome showing the locations of Lamp2, Atrx, and the X-inactivation center (Xic).
(D) Linear correlation between the ratio of the allele-specific cDNA amounts and the ratio of emitted fluorescence for each allele. cDNAs derived from homozygous B6 and Sp mice were mixed at five different ratios, and qRT-PCR was performed using TaqMan probes for the Lamp2 or Atrx gene. The square of the correlation coefficient is indicated by R2.
(E) Relative abundance of allele-specific transcripts in drug-selected MEFs. The relative abundances of allele-specific transcripts in HAT- or 6-TG-selected bsMEFs were calculated by using TaqMan probes for Lamp2 or Atrx.
Data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01. n.s., not significant.