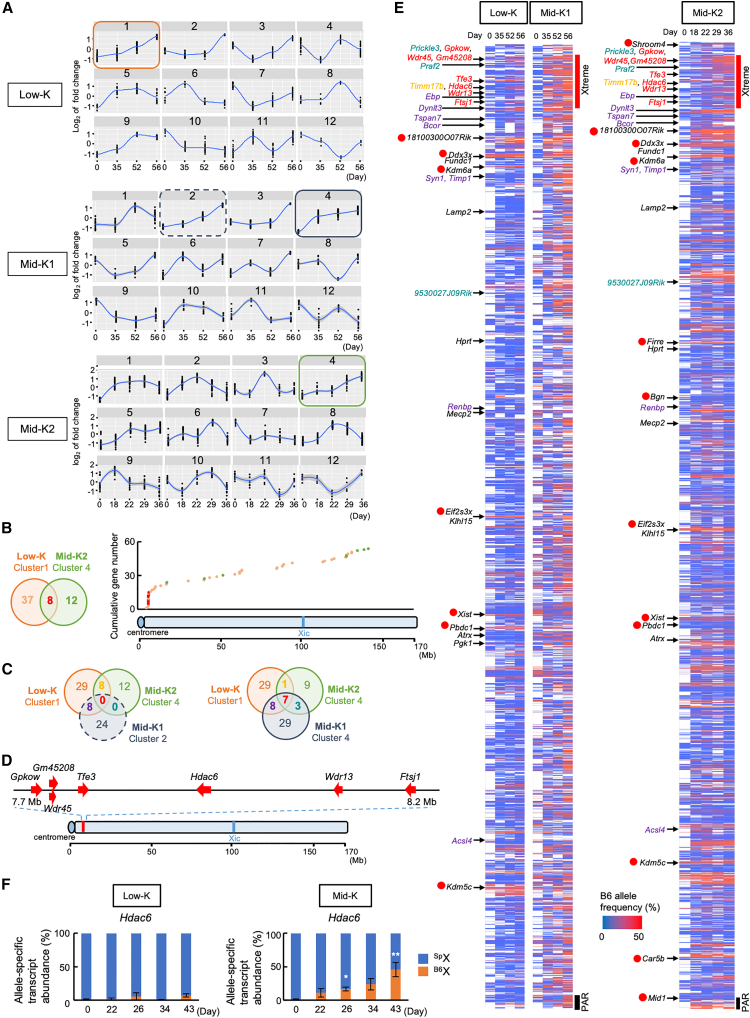
Figure 3.
Expression profiles of the genes on the Xi during reprogramming
(A) Clustering of expression patterns of genes reactivated on the Xi. X-linked genes were assigned into 12 clusters by expression patterns in Low-K, Mid-K1, or Mid-K2 cells. The colored squares indicate clusters that show a trajectory of increasing expression.
(B) X chromosomal locations of the genes that show increasing expression and are shared between Low-K and Mid-K2. A Venn diagram shows overlapping genes between Low-K cluster 1 and Mid-K2 cluster 4 (left panel), and a scatter diagram shows distribution of the all genes included in Low-K cluster 1 and Mid-K2 cluster 4 (right panel). The colors of dots in the scatter diagram reflect the ones in the Venn diagram.
(C) Venn diagrams show overlapping genes among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 2 (left) as well as those among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 4 (right).
(D) Locations of the seven genes that show early-onset reactivation.
(E) Heatmaps of transcriptional reactivation of the genes on the Xi chromosome. B6 allele frequencies in each polymorphic position were ordered by their genomic locations on the X chromosome. The genes that are common in among Low-K cluster 1, Mid-K2 cluster 4, and Mid-K1 cluster 4 are colored in yellow (one gene), red, (seven genes), purple (eight genes), and green (three genes), as in the right diagram in (C). Genes regarded as escapee genes are marked with red circles. Xtreme region and pseudoautosomal region (PAR) are shown with red and black lines, respectively.
(F) Relative abundances of allele-specific transcripts of Hdac6 in HAT-bsMEFs reprogrammed under the Low-K and Mid-K conditions. The same cells prepared in Figure 2B were analyzed.
Data represent mean ± SEM of at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01 versus HAT-bsMEFs (day 0).