Figure 6.
KDM1A occupancy in the Xtreme region
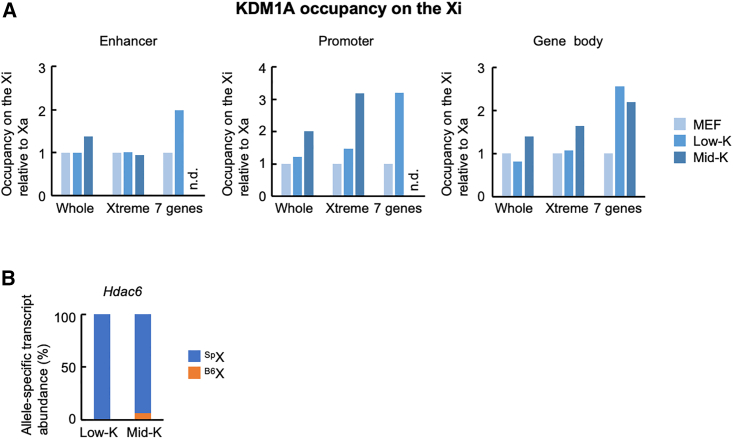
(A) KDM1A occupancy in different XCR states. HAT-bsMEFs were reprogrammed under the Low-K or Mid-K condition for about 20 days. The extracted chromatins were used for allele-specific ChIP-seq for KDM1A. KDM1A occupancies on the Xi relative to its occupancies on the Xa at enhancer, promoter, and gene body are shown. Relative KDM1A occupancies on the Xi in MEFs are set to 1. Seven genes are Gpkow, Wdr45, Gm45208, Tfe3, Hdac6, Wdr13, and Ftsj1 in the Xtreme region. n.d., not detected.
(B) Relative abundance of allele-specific Hdac6 transcripts in the cells used for ChIP-seq.
Data consist of a single experiment.