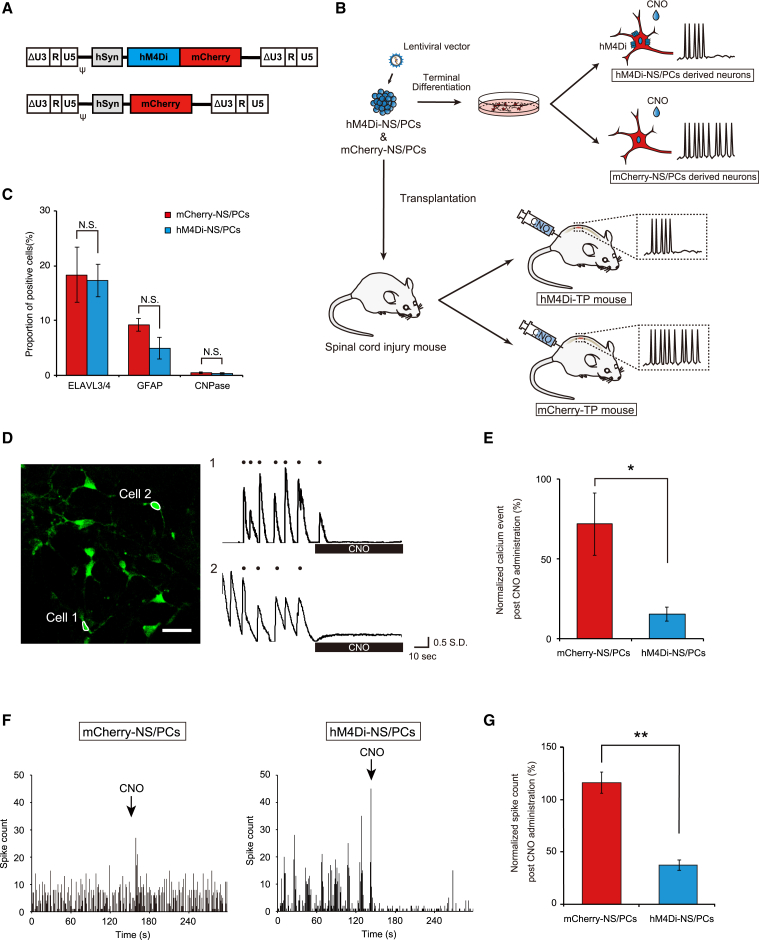
Figure 1.
Establishment of hM4Di-expressing NS/PCs; differential potential and inhibitory functional assessments in vitro
(A) Schematic of the integrated proviral form of the lentiviral vectors. Top: CSIV-hSyn-hM4Di-mCherry. Bottom: CSIV-hSyn-mCherry.
(B) Schematic of the experiments.
(C) Percentage of cell-type-specific marker cells in differentiated cells of mCherry-NS/PCs and hM4Di-NS/PCs (mCherry-NS/PCs, n = 4 wells/2 independent experiments; hM4Di-NS/PCs, n = 4 wells/2 independent experiments).
(D) Representative calcium transients of neurons differentiated from hM4Di-NS/PCs. Left: an image of neurons expressing jGCaMP7f in a single field of view. Scale bar, 30 μm. Right: time traces of representative cells highlighted with white outlines in the left image (the black dot indicates the calcium event).
(E) Average of normalized calcium events after CNO administration for neurons differentiated from mCherry-NS/PCs and hM4Di-NS/PCs (mCherry-NS/PCs, n = 15 cells/3 independent experiments; hM4Di-NS/PCs, n = 18 cells/3 independent experiments).
(F) Evaluation of neural activity by multi-electrode array assay. Representative time traces of the spike count by CNO administration (left, mCherry-NS/PCs; right, hM4Di-NS/PCs).
(G) Average normalized spike counts after CNO administration of neurons differentiated from mCherry-NS/PCs and hM4Di-NS/PCs (mCherry-NS/PCs, n = 10 wells/2 independent experiments; hM4Di-NS/PCs, n = 10 wells/2 independent experiments). ∗p < 0.05, ∗∗p < 0.01, and N.S., not significant according to the Mann-Whitney U test (C, E, and G). The data are presented as the mean ± SEM.