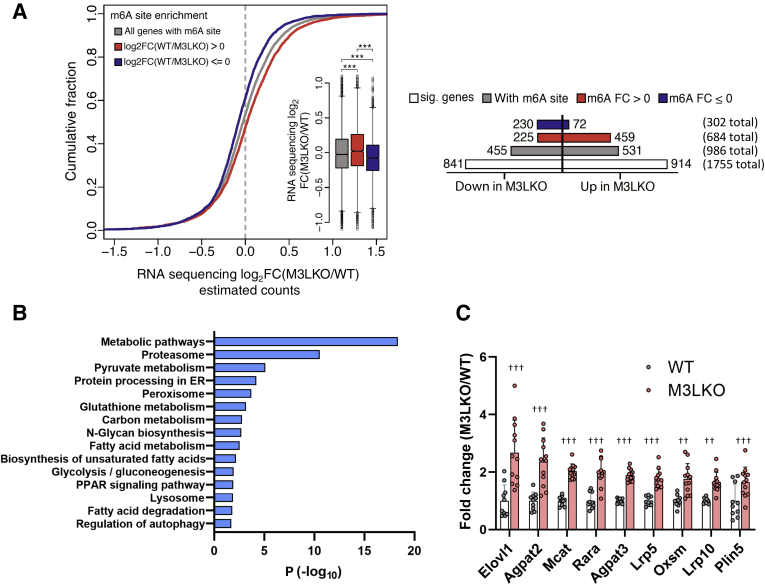
Figure 3.
RNA sequencing (RNA-seq) analysis reveals polyadenylated RNAs are differentially expressed in liver-specific METTL3 knockout (M3LKO) livers compared with wild-type (WT) mice livers. A: Cumulative fraction plot of poly A+ RNA expression in response to m6A enrichment (left panel). Transcripts were grouped in the following ways: those harboring m6A sites (gray), those with sites exhibiting reduced or lost m6A methylation [red; m6A enrichment log2 fold change (FC)(WT/M3LKO) > 0], and those with m6A sites with increased or unchanged methylation [blue; m6A enrichment log2FC(WT/M3LKO) ≤ 0] in M3LKO livers relative to the WT livers. Inset: Box plots depict differential poly A+ RNA abundance in these three groups classified based on m6A site enrichment. A horizontal bar plot (right panel) depicting the overall number of significantly altered transcripts (Welch test) in the following groups, from top to bottom, is presented: transcripts enriched or unaltered in M3LKO (m6A ≤ 0) (302 total; blue bar), transcripts harboring m6A sites lost in M3LKO livers (m6A > 0) (684 total; red bar), transcripts with m6A marks in both (986 total; gray bar), and all transcripts (with and without m6A) (1175 total; unfilled bar). B: The Kyoto Encyclopedia Gene and Genome pathway analysis of genes significantly dysregulated in M3LKO livers relative to WT livers in 5-week–old mice. C: RNA sequencing data demonstrating significant up-regulation of expression of select genes with m6A marks involved in metabolic pathways in M3LKO livers compared with WT livers (M3LKO/WT). n = 6 for WT (4 male and 2 female) mice; n = 8 M3LKO (4 male and 4 female) mice. ∗∗∗P < 0.001 (U-test); ††P < 0.01, †††P < 0.001 (Wald t-test). ER, estrogen receptor; PPAR, peroxisome proliferator-activated receptor; sig., significant.