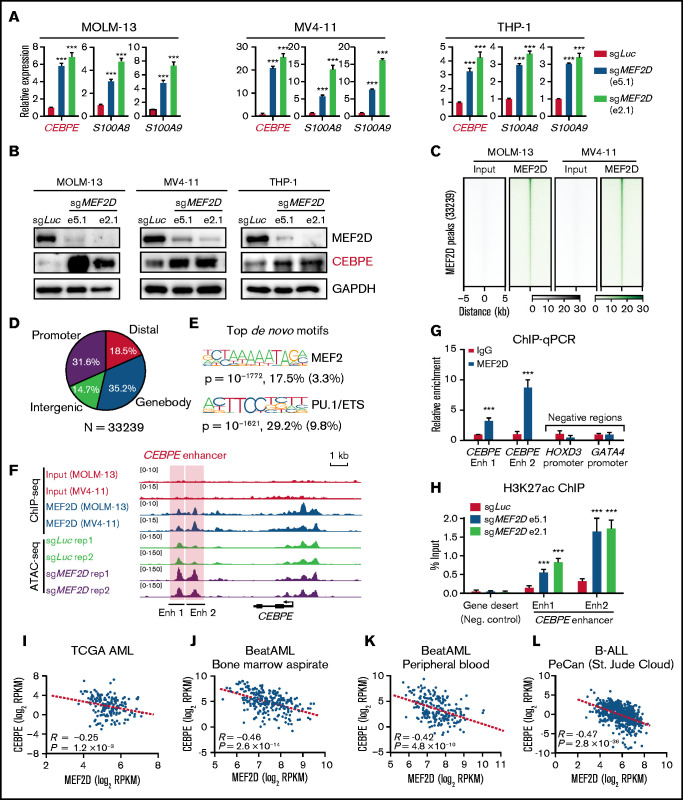
Figure 5.
MEF2D directly binds to CEBPE and negatively regulates CEBPE expression and enhancer histone acetylation. (A) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) showing expression of CEBPE and its target genes S100A8 and S100A9 in indicated AML cells with or without MEF2D knockout. (B) Western blot analysis of CEBPE in indicated control and MEF2D knockout AML cells. (C) Heatmap displaying MEF2D ChIP-seq read densities of 33 239 commonly identified from MOLM-13 and MV4-11 cells. (D) Genomic distribution of MEF2D ChIP-seq peaks commonly identified from MOLM-13 and MV4-11 cells. (E) Top de novo motifs identified from MEF2D ChIP-seq peaks. (F) ATAC-seq and MEF2D ChIP-seq profiles at the CEBPE locus in indicated leukemic cells. (G) ChIP-qPCR validating MEF2D binding at CEBPE enhancer regions. (H) ChIP-qPCR showing enrichment of H3K27ac at CEBPE enhancer regions. (I-K) CEBPE and MEF2D expression levels are negatively correlated in human AML patients. (L) CEBPE and MEF2D expression levels are negatively correlated in human B-ALL patients. ***P < .001.