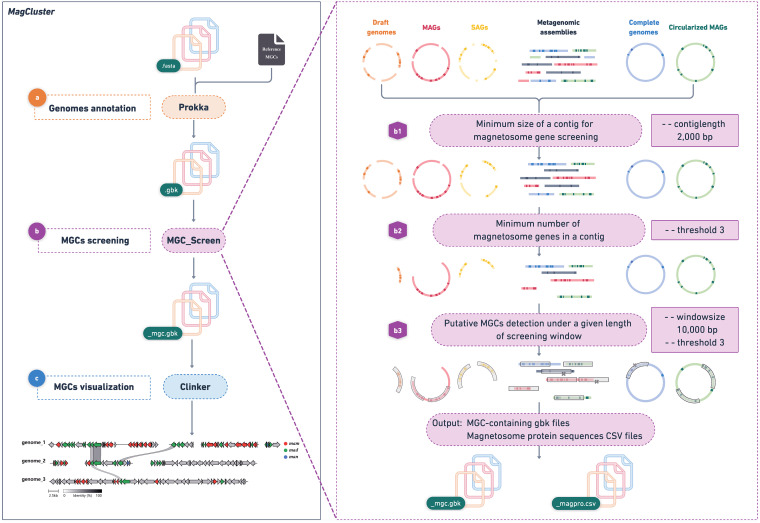
FIG 1.
Overview of the MagCluster workflow. (a) Genomes are annotated using Prokka with a mandatory reference file of magnetosome proteins via ‐‐proteins. (b) Putative MGCs or MGC-containing contigs are retrieved by the MGC_Screen module from GenBank files generated by the annotation module. First, contigs are filtered by the contig length (‐‐contiglength) and the minimum number of magnetosome genes in a contig (‐‐threshold). Then, the length of a genomic region containing no less than the given number of magnetosome genes is checked to meet the value of ‐‐windowsize. Finally, contigs that pass all restrictions are regarded as putative MGC-containing contigs. (b1) Contigs shorter than 2,000 bp (by default) are discarded. (b2) Magnetosome genes are identified through a text-mining strategy using the keyword “magnetosome” in protein names, and contigs containing fewer than 3 (by default) magnetosome genes are discarded. (b3) Putative MGCs are screened under a 10,000-bp (by default) window, and the minimum number of magnetosome genes (3 by default) in each window size is rechecked. (c) Putative MGCs are compared and visualized using clinker. MAGs, metagenome-assembled genomes; SAGs, single amplified genomes.