Abstract
Antibiotic collateral sensitivity (CS) occurs when a bacterium that acquires resistance to a treatment drug exhibits decreased resistance to a different drug. Here we identify reciprocal CS networks and candidate genes in Burkholderia multivorans. B. multivorans was evolved to become resistant to each of six antibiotics. The antibiogram of the evolved strain was compared to the immediate parental strain to determine CS and cross-resistance (CR). The evolution process was continued for each resistant strain. CS interactions were observed in 170 of 279 evolved strains. CS patterns grouped into two clusters based on the treatment drug being a beta-lactam antibiotic or not. Reciprocal pairs of CS antibiotics arose in at least 25% of all evolved strains. Sixty-eight evolved strains were subjected to whole-genome sequencing and the resulting mutation patterns were correlated with antibiograms. Analysis revealed there was no single gene responsible for collateral sensitivity (CS), and that CS seen in B. multivorans is likely due to a combination of specific and non-specific mutations. The frequency of reciprocal CS, and the degree to which resistance changed, suggests a long-term treatment strategy; when resistance to one drug occurs, switch to use of the other member of the reciprocal pair. This switching could theoretically be continued indefinitely, allowing life-long treatment of chronic infections with just two antibiotics.
Keywords: Collateral sensitivity, Antibiotic resistance, Burkholderia
1. Introduction
Burkholderia multivorans is a member of the Burkholderia cepacia complex (Bcc), a group of closely related Gram-negative bacterial species that are inherently resistant to many antibiotics. Several Bcc species can cause chronic and debilitating lung infections in cystic fibrosis patients [1], and after establishment, Bcc infections are usually chronic and eradication is difficult. Antibiotic therapy use in CF patients is common, both prophylactically and in response to pulmonary exacerbations, and this therapy creates an environment that selects for resistant mutants.
Bacterial antimicrobial susceptibility profiles change in response to selective pressure. Collateral changes in susceptibility arise when a mutant has an increase or decrease in resistance to an antibiotic to which the bacterium has not been exposed. Cross resistance (CR) is when a strain has increased resistance to a non-treatment drug, and collateral susceptibility (CS) is when a strain has decreased resistance to a non-treatment drug [2–4]. Experiments on Escherichia coli [2, 4, 5], Pseudomonas aeruginosa [6–8], and Staphylococcus aureus [9] have demonstrated cross resistance and collateral susceptibility. Reciprocal CS, which occurs when a pair of antibiotics switch between resistance and susceptible over lineage evolution, is less common but has greater clinical significance [2, 8].
Our laboratory has documented that collateral susceptibility (CS) and cross-resistance exists in a B. multivorans clinical isolate and occur in patterns based on the treatment drug [10]. We have previously reported on 13 independently evolved strains that exhibited CS and CR. Here, we further evolved 68 strains and document a pattern of reciprocal CS in B. multivorans. as well as report mutations acquired during selection which correlate with CS pairs. Reciprocal CS drug-pair combinations allow a treatment strategy of switching between two antibiotics instead of using a sequential therapy regiment that leads to pathogens with expanding antibiotic resistance.
2. Materials and Methods
2.1. Strains, culture conditions, and antibiotics
Strain AS149, a B. multivorans isolated from the sputum of a cystic fibrosis patient [11], as the ancestral parent. Bacteria were grown at 37° on LB for routine culturing and during experimental evolution and on Mueller-Hinton broth 2 (Sigma-Aldrich) for antimicrobial susceptibility testing.
Antibiotics involved in this study were chosen due to their inclusion in the CLSI [12] list of standard antibiotics tested against B. cepacia and for having varied targets, which we separated in βLA (meropenem [MEM], ceftazidime [CAZ]) and non-βLA (chloramphenicol [CHL], levofloxacin [LVX], minocycline [MIN], trimethoprim- sulfamethoxazole [SXT]) groups. BBL Sensi-Disc antimicrobial susceptibility test disks (BD) were used for all except minocycline (Oxoid). The minimum inhibitory concentration was determined using ETEST gradient strips.
2.2. Experimental evolution
Strains were evolved for resistance to one of six treatment drugs using a previously described method [13]. Evolved strains were classified as ‘resistant’ using CLSI zone of inhibition (ZOI) breakpoints or, for levofloxacin and chloramphenicol that lack disk diffusion breakpoints, when there was confluent growth up to the disk.
Lineages were created from AS149. A “lineage” includes all progeny that started with one treatment drug. Treatments used for all lineages is demonstrated for meropenem (MEM): AS149 is exposed to MEM until resistant (Progeny1MEM-R). Disk diffusion testing is performed using the treatment drug, MEM, and five non-treatment drugs to ensure the MEM-R strain is resistant to MEM and not a persister or cheater. If the MEM-R strain exhibits a change in the ZOI for any of the five non-treatment drugs, it was used to evolve a new strain for resistance to the non-treatment drug (e.g., CAZ-R [Progeny2CAZ-R]). Each evolved strain (e.g., CAZ-R) was then examined for changes in susceptibility to the five non-treatment drugs. If any collateral sensitivity interaction was observed, CAZ-R was subjected to the same treatment as was used with MEM-R to evolve progeny strains resistant to the CS non-treatment drugs. Reciprocal CS occurs if the Progeny2 strain loses resistance to the antibiotic used to select for the Progeny1 strain (i.e. if when Progeny1MEM-R is evolved to CAZ-R [i.e. becomes Progeny2CAZ-R] it simultaneously loses resistance to MEM). A lineage was terminated for one of two reasons: 1) the terminal strain had no CS or 2) reciprocal CS had been demonstrated twice.
2.3. Antimicrobial susceptibility testing and interpretation for collateral resistance and sensitivity
Antimicrobial susceptibility testing (AST) was performed as described previously [13]. The antibiogram of the evolved strain was compared to the antibiogram of the immediate parental strain to determine collateral sensitivity (CS) and cross-resistance (CR). Any change in zone of inhibition (ZOI) of 20% or greater for a non-treatment drug reflects collateral changes in susceptibility, with an increase in ZOI indicating CS and a decrease in ZOI indicating CR [10]. On strains having CS, an ETEST® was used to determine the minimum inhibitory concentration (MIC).
2.4. Statistical Analysis
All statistical analyses were performed with GraphPad Prism. An observed-versus-expected binomial one-tailed test was used to determine statistical significance of clustering in cross-resistance (CR) and collateral sensitivity (CS) regarding βLA and non-βLA clusters. Numbers of interactions, and not strains, were used since strains could contain more than one interaction. The null hypothesis was that any of the 5 non-treatment drugs had an equal chance to be the drug in the observed interaction. When the antibiotic was a βLA, expected values are 20% βLA (1 of 5) and 80% non-βLA (4 of 5); when a non-βLA, expected values are 40% βLA (2 of 5) and 60% non-βLA (3 of 5). Decreases in MICs are expressed as fold-changes for each non-treatment drug. Tests for normality (Anderson-Darling and Kolmogorov-Smirnov) were negative for all, so Kruskal-Wallis test and Dunn’s multiple comparisons test were run.
2.5. Genomic analysis
Sixty-eight independently evolved B. multivorans isolates, three parental AS149 biological replicates, and eight negative control biological replicates (2x five exposures, 10 exposures, 15 exposures, and 20 exposures) were subjected to whole genome sequencing (WGS). Negative control strains were constructed by sequentially plating one colony grown without antibiotic selection under the same procedure as evolved strains to account for mutations accumulated due to laboratory evolution. A single colony from each strain and two morphologically distinct colonies from control plates were sequenced by Omega Bioservices, using 151 bp paired end reads with an Illumina HiSeq 2500 platform. Variant filtering and analysis were performed as previously described [10].
To identify candidate genes likely involved in direct resistance and collateral sensitivity, starting with the evolved strains of interest, we first eliminated mutations from the ancestral parent, AS149, and those in the negative control strains. Mutations shared between pairs of reciprocal CS strains and mutations greater than 100 bp upstream of a downstream gene in intergenic regions were not accounted for in this analysis. Predicted protein loss of function mutations within structural genes were designated as high impact.
3. Results
3.1. Frequency of cross-resistance and collateral susceptibility
Of 279 evolved strains, 188 (67%) exhibited cross-resistance and 170 (61%) exhibited collateral susceptibility. The frequency for each of the six antibiotics is seen in Table 1. As was observed previously [10], interaction patterns were observed based upon the treatment drug; the beta lactam antibiotics (βLA) forming one cluster (134 strains [48%]) and the non-beta lactam antibiotics (non-βLA) forming the other (145 strains [52%]).
Table 1.
Strains evolved and collateral changes observed.
Results of experimental evolution are shown by treatment drugs: chloramphenicol (CHL), ceftazidime (CAZ), levofloxacin (LVX), meropenem (MEM), minocycline (MIN), and trimethoprim-sulfamethoxazole (SXT). Information for each treatment drug includes the total number of strains evolved, the percentage of strains showing cross-resistance (CR) and collateral sensitivity (CS), and the average number and range of CS interactions per CS-exhibiting strain.
| Treatment Drug | Total # strains evolved | Percentage with CR | Percentage with CS | Avg # CS interactions per strain w/CS | Range of CS interactions per strain |
|---|---|---|---|---|---|
| CHL | 20 | 90% | 65% | 1.6 | 1–3 |
| CAZ | 62 | 65% | 56% | 1.7 | 1–4 |
| LVX | 44 | 75% | 59% | 1.3 | 1–3 |
| MEM | 72 | 58% | 61% | 2.1 | 1–5 |
| MIN | 34 | 71% | 56% | 1.3 | 1–3 |
| SXT | 47 | 66% | 70% | 2.0 | 1–5 |
| TOTALS | 279 | 67% | 61% | 1.8 | 1–5 |
3.2. Cross-resistance patterns
The percentage of evolved strains exhibiting CR as well as the average CR interactions per strain were determined and compared for each of the six antibiotics being used as the treatment drug. The ratio of CR-exhibiting strains to total evolved strains for both βLA resistance groups was lower (61%, 82 of 134) than for the four non-βLA resistance groups (73%, 106 with CR of 145 evolved). For all 188 CR-exhibiting strains, there as an average of 1.9 CR interactions per strain. If treatment drugs are separated into βLA and non-βLA clusters, the range for n-βLAs is 1.7–2.3 and for βLA is 1.7–1.8.
Most cross-resistance interactions were observed within clusters (Fig. 1A). When the treatment drug was a non-βLA, any CR interaction observed was more likely to be with another non-βLA. The same pattern is seen in the βLA cluster; >70% of CR-exhibiting strains with a βLA treatment drug had CR interactions occur in the other βLA. Between-cluster interactions were statistically more common when the treatment drug was a βLA than a non-βLA (see Fig. S1A). We performed an observed-versus-expected, one-tailed binomial statistical test; clustering was statistically significant for all treatment drugs.
Figure 1.
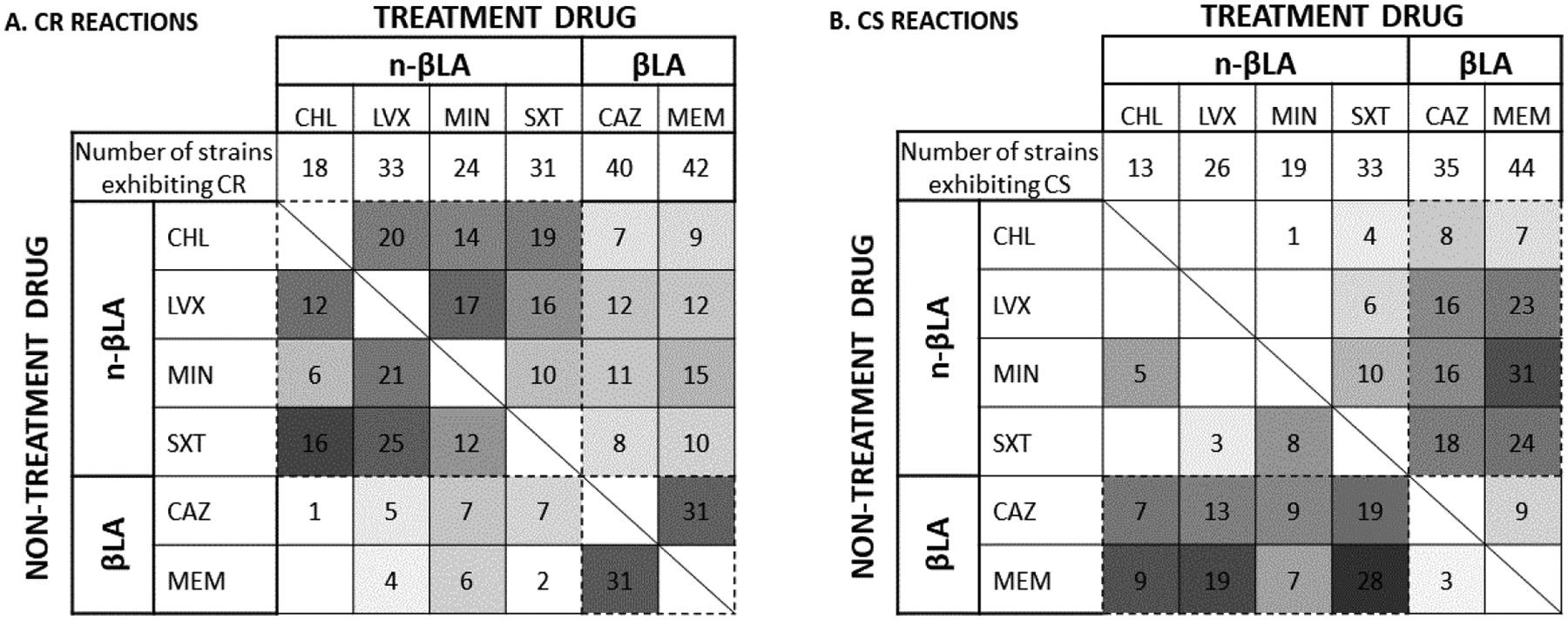
Heat map of cross-resistance (CR) and collateral sensitivity (CS) interactions
For each treatment drug - chloramphenicol (CHL), ceftazidime (CAZ), levofloxacin (LVX), meropenem (MEM), minocycline (MIN), and trimethoprim-sulfamethoxazole (SXT) - used to evolve a strain (listed across the top) and the non-treatment drug to which the strain exhibits a change in sensitivity (on the left), the number of strains that demonstrated cross-resistance (Panel A) or cross-sensitivity (Panel B) is indicated in each box. The percentage of evolved strains exhibiting the reaction to each non-treatment drug are represented by the intensity of the cell. Groupings within the βLA and non-βLA clusters are bordered by dashed lines.
3.3. Collateral susceptibility patterns
Of the 170 strains with collateral sensitivity (CS), the majority had CS in one or two non-treatment drugs (NTD) with an average number of CS interactions per strain of 1.8 (Table 1): 45% had increased sensitivity to only one non-treatment drug (NTD) and 41% increased sensitivity to two NTDs. Meropenem had the highest average of CS interactions per strain (2.1) and levofloxacin and minocycline had the lowest (1.3). Two groups, trimethoprim-sulfamethoxazole and meropenem, had strains with 5 interactions, i.e. all non-treatment drugs demonstrated CS. The absolute ratio of CS-exhibiting strains to total evolved strains did not significantly differ between these two clusters (59% for the βLA and 63% for the non-βLA).
The clusters identified from cross-resistance interaction patterns (βLA, non-βLA) were also present in collateral sensitivity patterns. Unlike CR interactions that were within clusters, most CS interactions were between clusters (Fig. 1B and Fig. S1B). Clustering was statistically significant for all treatment drugs.
3.4. Quantitative decrease in MIC for collateral sensitivity interactions
The degree to which an evolved strain changed resistance to a non-treatment drug was measured by comparing minimum inhibitory concentration (MIC) values obtained using ETEST for all non-treatment drugs. Ceftazidime showed the greatest decrease, with an average fold decrease of 86.1 (SD 204.2, range 1.3–1024); chloramphenicol (mean 2.9, SD 2.2, range 1.3–8) and levofloxacin (mean 2.8, SD 2.0, range 1.3–12) had the smallest decreases.
3.5. Reciprocal CS between antibiotic pairs
Identification of two treatment drugs which lead to collateral sensitivity in the other antibiotic, i.e. reciprocal CS, suggests a strategy to treat chronic infections. Candidate reciprocal CS pairs were selected as follows: each antibiotic of the pair has a relatively high percentage of strains with collateral sensitivity as the non-treatment drug when the other was the treatment drug. Pairs were ranked by their average, combined CS percentage. The top five pairs are given with the percentage of strains resistant to the treatment drug exhibiting CS to the other antibiotic in the pair presented in the subsequent parentheses. They are: trimethoprim-sulfamethoxazole (SXT) / meropenem (MEM) (non-treatment drug = MEM 60% [i.e. 60% of strains resistant to SXT show CS to MEM], SXT 33%, average combined CS 46%), LVX/MEM (43, 32, 38), SXT/CAZ (40, 29, 35), minocycline (MIN)/MEM (21, 43, 32), and LVX/CAZ (30, 26, 28). A diagram of these pairings is given in Fig. S2.
3.6. Genes involved in collateral resistance and reciprocal collateral sensitivity
Whole genome sequencing and mutational analysis were conducted on the parental and 68 of the evolved strains. The average number of mutations per strain were 50.2 (range: 13–120); 23 (range: 1–67) intergenic, and for coding regions, there were an average of 13.9 (range: 2–31) synonymous and 13.3 (range: 4–25) nonsynonymous mutations.
Genetic analysis focused on two reciprocal pairs: MEM-LVX and CAZ-LVX (Table S1, S2). Because MEM and CAZ have similar mechanisms of action and both show reciprocal CS with LVX, comparing mutations acquired between these two pairs should reveal those most likely to be involved in CS.
Isolates evolved to treatment drug LVX with increased susceptibility to MEM acquired unique mutations within six structural genes and two intergenic regions (Table S3). Of the six structural genes, three genes acquired high impact mutations (rseP, dacB, and fimV). Isolates evolved to treatment drug MEM with increased susceptibility to LVX acquired unique mutations within nine structural genes and nine intergenic regions. Three structural gene mutations were designated as high impact (rimO, snoaL-like polyketide cyclase, and rhaT).
Isolates evolved to treatment drug LVX with increased susceptibility to CAZ acquired unique mutations within eight structural genes and two intergenic regions (Table S4). Of the eight structural genes, three acquired high impact mutations (bpeR, fimV, phage lysozyme). Isolates evolved to treatment drug CAZ with increased susceptibility to LVX acquired unique mutations within eleven structural genes and seven intergenic regions. Of the eleven structural genes, five acquired high impact mutations (rne, tRNA-ser, dacB, fabI, and a transmembrane protein).
4. Discussion
This study identifies and quantitates reciprocal collateral sensitivity (CS) antibiotic pairs in B. multivorans, and identifies candidate genes involved in CS. The frequency of collateral susceptibility changes may be taxa-specific as what we observed was higher [14] and lower [2] than found previously with other organisms.
From the six antibiotics used in this study, representing multiple modes of action, collateral sensitivity (CS) patterns grouped into two clusters based on the treatment drug being a beta-lactam antibiotic (βLA) or not (non-βLA). The majority of cross-resistance (CR) interactions are within each cluster, consistent with a previous report [10]. Here we also observed more between-cluster CR interactions when the treatment was a βLA than when a non-βLA (Fig. S1A). Collateral sensitivity interactions were observed in 170 of 279 evolved strains, with the majority (55%) having more than one interaction. When present, these interactions were usually seen between clusters (Fig. 1B and S1B), with the sole exception being when minocycline is the treatment drug.
The relevance of collateral sensitivity (CS) to clinical use is influenced by the degree to which there is a collateral decrease in resistance and how reproducible are reciprocal CS pairs. When analyzing the degree of change in the MIC observed with collateral sensitivity, we noted that seven strains (12%) exhibited a decrease of greater than 100-fold, and almost half (49%, 28 strains) exhibited a decrease of >25-fold (data not shown). The highest change in MIC was observed with ceftazidime, with a 1024-fold decrease. For all other non-treatment drugs, the average fold decrease was 7.1 with a standard deviation of 11.8 (data not shown). Reciprocal CS pairs arose in at least 25% of all evolved strains. The pairs having the highest CS frequency, and hence of most clinical use, include MEM-SXT, MEM-LVX, CAZ-SXT, MEM-MIN, and CAZ-LVX (Fig. S2).
Analysis of whole genome sequencing data revealed there was no single gene responsible for collateral sensitivity (CS), but instead is likely due to a combination of specific and non-specific mutations. Collateral sensitivity that we observed resulted from a combination of mutations in specific antibiotic targets and non-specific processes, polar effects from operon mutations, and mutations within efflux pump regulators [15,16].
Mutations in specific antibiotic targets, such as penicillin binding protein PBP1a and class A beta-lactamase bla, were observed in both MEM and MEM/CAZ evolved isolates; respectively [17,18]. Isolates resistant to MEM acquired either a large disruptive insertion within PBP1a or missense mutations resulting in a BlaN145S substitution. Isolates that acquired a BlaD192G substitution alternatively showed an increase in susceptibility to MEM. In addition, isolated resistant to CAZ acquired a BlaE179G substitution (Fig. 2). This phenomenon was observed across diverse reciprocal pairs alluding to the position of amino acid substitution within a beta-lactamase playing a role in altering antibiotic specificity and resistance phenotype.
Figure 2:
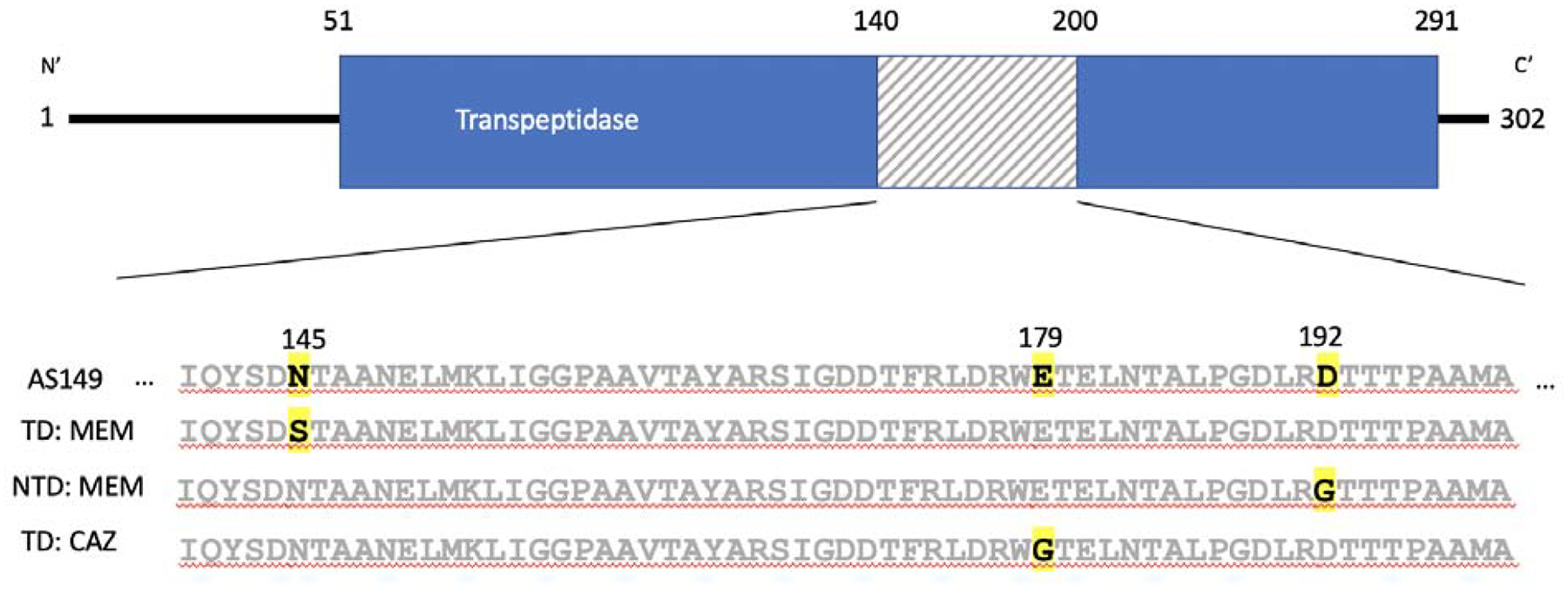
Schematic of Bla protein (NP80_4859). Bla1–51 is a signal peptide sequence at the N’ terminus followed by the main enzymatic region of the beta lactamase, Bla51–291, represented in blue and the C’ terminus, Bla291–302. The region in the protein where mutations were localized in CAZ and MEM CS are represented in the shaded box. This region’s amino acid sequence for parental strain, AS149, and representative sequences for strains exhibiting CS is given below. The TD or NTD is specified next to the sequence and amino acid change is bolded and highlighted for respective strain.
Non-specific mutations affecting membrane permeability were observed in LVX-MEM and CAZ-LVX reciprocal pairs. Resistance to LVX can be associated with one frameshift and three small deletions at the C’ terminus of the LPS heptosyltransferase (RfaF) protein affecting outer membrane porin concentration [19–21]. Isolates with increased susceptibility to MEM acquired a loss of function mutation in σE positive regulator, rseP. Previous work has shown σE plays a critical role in protecting Burkholderia from entry of meropenem [22].
ParB and known fluroquinolone target, ParC, are co-transcribed on the same operon, parCBA [23]. Mutations within parB may have polar effects on parC activity, however the consequence of mutations in parB and fluroquinolone activity have yet to be investigated. Altering mutations within bpeT, the regulator of fluroquinolone efflux pump BpeEF-OprC, determined the resistance (BpeTGlu177Gly) or susceptibility (BpeTGlu211Gly) to LVX in LVX-CAZ reciprocal pair. A similar pattern was observed in B. pseudomallei bpeT [24,25].
All isolates acquired mutations that affect biofilm formation and/or motility (fimV or flgL) and mutations in genes that are associated with resistance to antibiotics not included in this study. For example, aminoglycosides (rimO, hflK, bpeR) [26–28], penicillin and monobactams (dacB) [29], sulfamethoxazole (purU) [30], vancomycin (lamB) [31], and fatty acid inhibitors (fabI) [32]. A potential bet-hedging strategy under continued selection for antibiotic resistance during evolution of the lineages.
Collateral sensitivity (CS) patterns in E. coli and P. aeruginosa have previously been shown to differ based upon ancillary mutations that arise during prior antibiotic exposure [8, 33, 34]. The model is that mutations that arose during previous antibiotic exposure did not change the antibiotic resistance profile, but ameliorated the effect of subsequent mutations on antibiotic resistance. This pattern is validated by the reoccurrence of mutated genes in this extended study that were seen in the 13 first generation CS isolates [10].
Here we document the existence of multiple reciprocal CS pairs across numerous evolved generations in Burkholderia suggesting a long term treatment strategy; when resistance to one drug occurs, switch to use the other member of the reciprocal pair. If resistance to the second drug occurs, switch back to the initial class of drug. Further, we present genomic data analysis that identified candidate CS genes, highlighting potential mechanisms of antibiotic resistance and antibiotic susceptibility in B. multivorans.
Supplementary Material
Highlights.
Burkholderia exhibits reciprocal antibiotic collateral sensitivity (CS)
Candidate genes involved in CS are identified
Reciprocal CS networks can be used to guide treatment of chronic infections
Funding:
This work was supported by funds provided by The University of North Carolina at Charlotte and a National Institutes of Health grant 1R15HL126122-01 to T.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: None declared.
Ethical Approval: Note required.
References
- [1].Lipuma J The changing microbial epidemiology in cystic fibrosis. Clinical Microbiology Reviews 2010; 23:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Science Translational Medicine 2013; 5:204ra132–204ra132. [DOI] [PubMed] [Google Scholar]
- [3].Sass A, Marchbank A, Tullis E, LiPuma JJ, Mahenthiralingam E. Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 2011; 12:373–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lázár V, Nagy I, Spohn R, Csörgő B, Györkei Á, Nyerges Á, Horváth B, Vörös A, Busa-Fekete R, Hrtyan M, Bogos B, Méhi O, Fekete G, Szappanos B, Kégl B, Papp B, Pál C. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nature communications 2014; 5:4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Podnecky NL, Aarag E, Kloos JM, Sørum V, Primicerio R, Roberts AP, Rozen DE, Samuelsen Ø, Johnsen PJ. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nature Communications 2018; 9:3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jansen G, Mahrt N, Tueffers L, Barbosa C, Harjes M, Adolph G, Friedrichs A, Krenz-Weinreich A, Rosenstiel P, Schulenburg H. Association between clinical antibiotic resistance and susceptibility of Pseudomonas in the cystic fibrosis lung. Evolution, Medicine, and Public Health 2016:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Imamovic L, Ellabaan MMH, Dantas Machado AM, Citterio L, Wulff T, Molin S, Krogh Johansen H, Sommer MOA. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 2018; 172:121–134.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Roemhild R, Barbosa C, Beardmore RE, Jansen G, Schulenburg H. Temporal variation in antibiotic environments slows down resistance evolution in pathogenic Pseudomonas aeruginosa. Evolutionary Applications 2015; 8:945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rodriguez de Evgrafov M, Gumpert H, Munck C, Thomsen TT, Sommer MOA. Collateral resistance and sensitivity modulate evolution of high-level resistance to drug combination treatment in Staphylococcus aureus. Molecular Biology and Evolution 2015;.32:1175–1185. [DOI] [PubMed] [Google Scholar]
- [10].Flanagan JN, Kavanaugh L, Steck TR. Burkholderia multivorans exhibits antibiotic collateral sensitivity. Microbial Drug Resistance. Published online August 8, 2019; DOI: 10.1089/mdr.2019.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stokell JR, Gharaibeh RZ, Steck TR. Rapid emergence of a ceftazidime-resistant Burkholderia multivorans strain in a Cystic Fibrosis patient. Journal of Cystic Fibrosis 2013; 12:812–816. [DOI] [PubMed] [Google Scholar]
- [12].CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. Document M100-S24 CLSI, Wayne, PA. [Google Scholar]
- [13].Flanagan JN, Steck TR. Use of antibiotic disks to evolve drug-resistant bacteria. Antonie van Leeuwenhoek 2018; 111:1719–1722. [DOI] [PubMed] [Google Scholar]
- [14].Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, Hrtyan M, Busa‐Fekete R, Bogos B, Méhi O, Csörgő B, Pósfai G, Fekete G, Szappanos B, Kégl B, Papp B, Pál C. Bacterial evolution of antibiotic hypersensitivity. Molecular Systems Biology 2013; 9:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anuwat A, Albert S, Mathias W, Wipa S. Porin involvement in cephalosporin and carbapenem resistance of Burkholderia pseudomallei. PLoS ONE 2014; 9:e95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Narayanaswamy V, Giatpaiboon S, Baker S, Wiesmann W, Lipuma J, Townsend S. Novel glycopolymer sensitizes Burkholderia cepacia complex isolates from cystic fibrosis patients to tobramycin and meropenem. PLoS One 2017; 12:e0179776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Siriyong T, Murray RM, Bidgood LE, Young SA, Wright F, Parcell BJ, Voravuthikunchai SP, and Coote PJ Dual B-lactam combination therapy for multi-drug resistant Pseudomonas aeruginosa infection: enhanced efficacy in vivo and comparison with monotherapies of penicillin-binding protein inhibition. Sci Rep 2019; 9(1):9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sarovich DS, Price EP, Von Schulze AT, et al. Characterization of Ceftazidime Resistance Mechanisms In Clinical Isolates of Burkholderia pseudomallei from Australia. PlosOne 2012; 7(2):e30789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Linkevicius M, Sandegren L, and Andersson DI Mechanisms and fitness costs of tigecycline resistance in E. coli. Journal of Antimicrobial Chemotherapy 2013; 68(12): 2809–2819. [DOI] [PubMed] [Google Scholar]
- [20].Aldred KJ, Kerns RJ, and Osheroff N Mechanism of Quinolone Action and Resistance. Biochemistry 2014; 53(10):1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grizot S, Salem M, Vongsouthi V, Durand L, Moreau F, Dohi H, Vincent S, Escaich S, Ducruix A. Structure of the Escherichia coli heptosyltransferase WaaC: binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. J Mol Biol. 2006; 363(2):383–94. [DOI] [PubMed] [Google Scholar]
- [22].Held K, Gasper J, Morgan S, Siehnel R, Singh P, and Manoil C Determinants of Extreme B-lactam tolerance in the Burkholderia pseudomallei complex. Antimicrobial Agents and Chemotherapy 2018; 62:e00068–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bagel S, Hullen V, Wiedemann B, and Heisig P Impact of gyrA and parC Mutations on Quinolone Resistance, Doubling Time, and Supercoiling Degree of Escherichia coli. Antimicrobial Agents and Chemotherapy 1999; 43(4):868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Podnecky NL, Rhodes KA, and Schweizer HP Efflux pump-mediated drug resistance in Burkholderia. Frontiers in Microbiology 2015; 6:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, et al. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS ONE 2012; 7:e36507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Carr JF, Hamburg D, Gregory ST, Limbach GP, and Dahlberg AE Effects of Streptomycin Resistance Mutations on Posttranslational Modification of Ribosomal Protein S12. Journal of Bacteriology 2006; 188(5):2020–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hinz A, Lee S, Jacoby K, and Manoil C Membrane Proteases and Aminoglycoside Antibiotic Resistance. Journal of Bacteriology 2011; 193(18): 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chan YY, Tan TM, Ong YM, Chua KL. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother. 2004;48(4):1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Moya B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, and Oliver A Pan-β-Lactam Resistance Development in Pseudomonas aeruginosa Clinical Strains: Molecular Mechanisms, Penicillin-Binding Protein Profiles, and Binding Affinities. Antimicrobial Agents and Chemotherapy 2012; 56(9): 4771–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vendantam G, Guay GG, Austria NE, Doktor SZ, and Nichols BP Characterization of Mutations Contributing to Sulfathiazole Resistance in Escherichia coli. Antimicrobial Agents and Chemotherapy 1998; 42(1):88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Choi U and Lee C Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli. Frontiers in Microbiology 2019; 10:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hoang TT and Schweizer HP Characterization of Pseudomonas aeruginosa Enoyl-Acyl Protein Reductase (FabI): a Target for the Antimicrobial Triclosan and Its Role in Acylated Homoserine Lactone Synthesis. Journal of Bacteriology 1999; 5489–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nichol D, Rutter J, Bryant C, Hujer AM, Lek S, Adams MD, Jeavons P, Anderson ARA, Bonomo RA, Scott JG. Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nature Communications 2019; 10:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gifford DR, Furio V, Papkou A, Vogwill T, Oliver A, MacLean RC. Identifying and exploiting genes that potentiate the evolution of antibiotic resistance. Nature Ecology & Evolution 2018; 2:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


