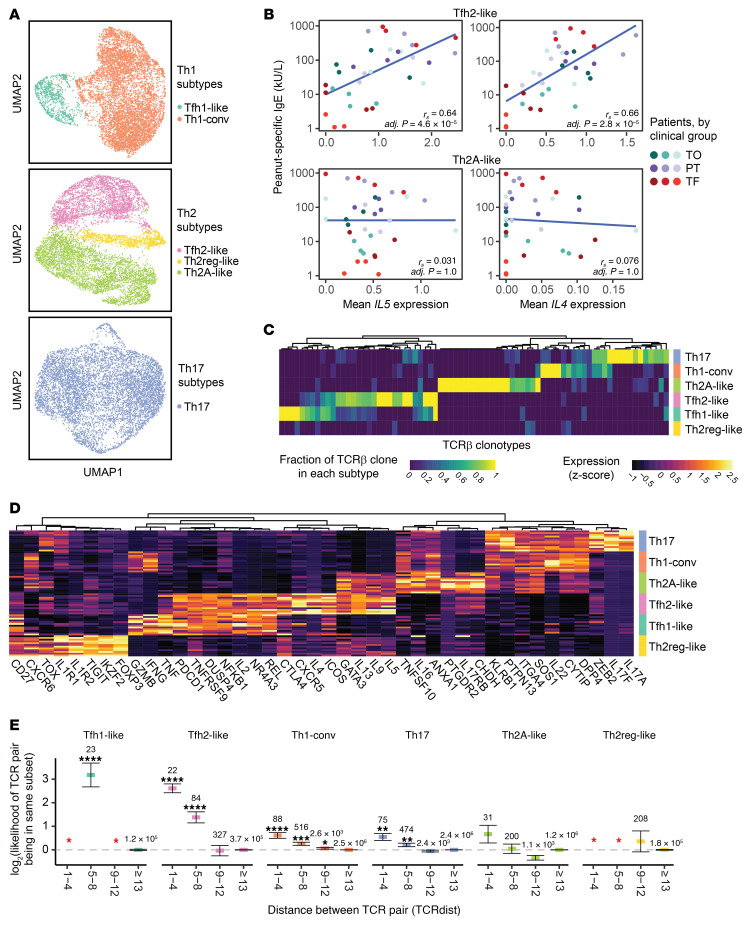
Figure 4. Peanut-reactive Th subtypes are clonally distinct and exhibit TCR convergence.
(A) UMAP visualizations of Th1- (n = 7,609 cells), Th2- (n = 7,877 cells), and Th17-scoring cells (n = 7111 cells). Clusters are annotated by their putative identity. (B) Scatter plots of the average expression of IL5 and IL4 in Tfh2-like cells or Th2A-like cells (for each patient at each time point) and peanut-specific IgE titers. Linear fit, Spearman’s correlation (rs, n = 34), and adjusted P values are shown. (C) Fraction of TCRβ clonotypes belonging to each subset. The fraction is defined as the number of cells of a TCRβ CDR3 sequence (column) detected in each Th subset, divided by the total number of cells within the clonotype. Clonotypes were randomly downsampled to visualize a comparable number from each subset. (D) Differentially expressed genes in each Th subset. Genes were selected using an ROC test and manual curation. Each row represents the scaled average gene expression in 1 patient. (E) TCR distance analysis of TCR sequences. The x axis represents bins of increasing pairwise TCR distance, calculated using TCRdist, and the y axis represents the likelihood of pairs of cells at a given TCR distance to be of the same Th subset, normalized to the prior probability of any 2 cells belonging to that subset (see Methods). ****P < 0.0001 (adjusted), ***P < 0.001 (adjusted), and **P < 0.01 (adjusted), by 2-sided χ2 proportion test with 1 degree of freedom. The total number of pairs within each TCR distance and subset is indicated above each data point. The red asterisk indicates that no pair of TCR sequences with the respective TCR distance bin was found in the respective subset. Error bars represent 85% binomial CIs. Data were combined from all patients at all time points (A–E).