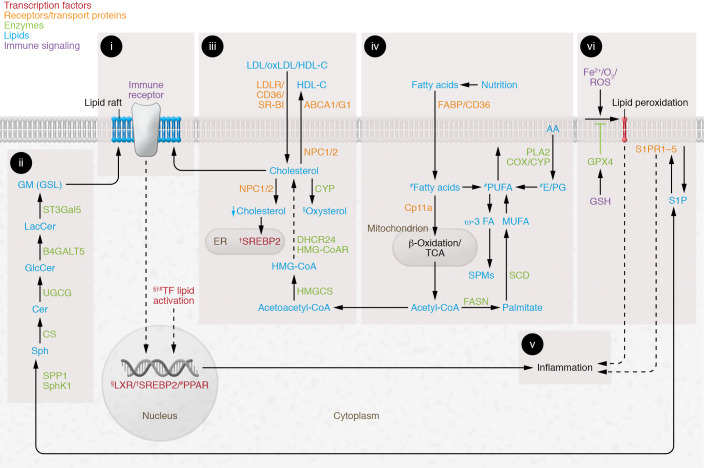
Figure 1. Summary of lipid metabolism pathways important for inflammation.
(i) Lipid rafts: cholesterol- and glycosphingolipid (GSL)-enriched cell signaling platforms. (ii) De novo GSL biosynthesis: differential expression influences immune receptor–mediated signaling and cell function. (iii) Intracellular cholesterol is regulated by liver X receptor (LXR) and sterol regulatory element–binding protein 2 (SREBP-2) (see v). LXR activation by oxysterols induces cholesterol efflux (ABCA1, ABCG1) and reduces lipid uptake (LDLR, VLDLR, SR-BI, CD36). Niemann-Pick type C1 (NPC1) and NPC2 regulate lysosomal/late endosomal trafficking/recycling of intracellular lipids. SREBP-2 opposes LXR and promotes cholesterol biosynthesis and uptake (HMGCoAR, LDLR). (iv) Nutrition influences fatty acid composition and metabolism. Fatty acids are metabolized to produce energy (ATP) by mitochondrial β-oxidation and TCA. Monounsaturated fatty acids (MUFAs) are synthesized via acetyl-CoA, fatty acid synthase (FASN), and stearoyl-CoA desaturase (SCD). Polyunsaturated fatty acids (PUFAs), diet-derived or biosynthesized in vivo, influence arachidonic acid (AA) metabolism. PUFAs are precursors to triglycerides, phospholipids in plasma membrane, second messengers, hormones, and ketone bodies. Prostaglandins (PGs) are produced following AA release from membrane phospholipids. Downstream PG signaling and eicosanoids have direct metabolic effects on immune cells via PPARs, mediating antiinflammatory effects and modulating LXRs (see v). Omega-3 (ω-3) PUFAs are enzymatically converted to antiinflammatory resolvins (specialized pro-resolving mediators [SPMs]). (v) Lipid metabolism activates transcription factors and influences inflammation via multiple mechanisms (see text). (vi) PUFA phospholipid peroxidation is induced by iron overload and ROS. Products of lipid peroxidation are eliminated via glutathione peroxidase-4 (GPX4); clearance defects induce cell membrane damage and ferroptosis. Sphingosine 1-phosphate (S1P) is derived from membrane phospholipids; activation via binding to S1P receptors (S1PR1–5) initiates immune cell localization to inflammatory sites and T cell differentiation. §, †, and # indicate processes activating LXR, SREBP2, or PPARs, respectively. Other abbreviations: B4GALT5, β-1,4-galactosyltransferase-5; Cer, ceramide; CS, ceramide synthase; DHCR24, 24-dehydrocholesterol reductase; FABPs, fatty acid–binding proteins; GlcCer, glucosylceramide; GM, GM1 ganglioside; GSH, glutathione; HMGCS, 3-hydroxy-3-methylglutaryl–CoA synthase-1; LacCer, lactosylceramide; ox, oxidized; Sph, sphingosine; SphK1, sphingosine kinase-1; SPP1, S1P phosphohydrolase-1; SR-BI, scavenger receptor class B type I; ST3Gal5, ST3 β-galactoside α-2,3-sialyltransferase-5; UGCG, UDP-glucose ceramide glucosyltransferase.