Abstract
We developed a single-tube rapid method for the detection and differentiation of varicella-zoster virus (VZV) vaccine and wild-type strains that combines rapid-cycle PCR with wild-type-specific fluorescent probe melting profiles for product genotyping. A region including the polymorphic site in VZV open reading frame (ORF) 62 was amplified in the presence of two fluorescence-labeled hybridization probes. During the annealing step of the thermal cycling, both probes bound to their complementary sequences in the amplicon, resulting in resonance energy transfer, thus providing real-time fluorescence monitoring of PCR. Continuous acquisition of fluorescence data during a melting curve analysis at the completion of PCR revealed that loss of fluorescence occurred in a strain-specific manner as the detection probe, which was fully complementary to the wild-type VZV ORF 62 region, melted off the template. Use of this method allowed genotyping of samples within minutes after the completion of PCR, eliminating the need for post-PCR sample manipulation. In addition to reducing the time required to produce a result, this method substantially reduces the risk of contamination of the final product as well as the risk of sample tracking errors. The genotypes of 79 VZV-positive samples determined by this fluorescent resonance energy transfer (FRET) method were identical to the genotypes obtained by conventional PCR and restriction fragment length polymorphism analysis. The genotyping of VZV strains by the FRET method is a rapid and reliable method that is suitable for typing and that is also practical for use for the processing of large numbers of specimens.
Varicella-zoster virus (VZV) is the etiologic agent of primary varicella (chicken pox) in childhood, establishing a latent infection that may reactivate to cause herpes zoster (shingles). VZV infections are usually benign, but serious and occasionally fatal infections do occur (4, 13, 24). Before an effective vaccine (Oka) was licensed, about 100 VZV-related deaths and 10,000 to 12,000 hospitalizations occurred annually in the United States (1, 2, 30, 31). The Oka vaccine protects most recipients (4, 6, 8, 27, 28, 30–32, 34), but mild breakthrough infections have been documented (5, 7, 28, 32, 34).
Localized or disseminated rashes sometimes develop within a few weeks after immunization. Rarely, secondary transmission of the Oka vaccine strain from vaccinees has occurred (18, 28). The vaccine strain occasionally reactivates to cause zoster (7, 9, 19).
Methods that reliably distinguish vaccine VZV strains from wild-type strains are needed to effectively monitor vaccine-related adverse events. Such methods are crucial for studies of duration of immunity and breakthrough infections and for analysis of VZV outbreaks. PCR amplification of selected VZV DNA sequences, followed by restriction enzyme digestion for detection of sequence variations, is a sensitive and reliable approach (10, 12, 16, 17, 22). VZV DNA can be amplified from vesicular fluid, scabs, papular scrapings, peripheral blood lymphocytes, and cerebrospinal fluid (15–17, 26), and PCR combined with restriction fragment length polymorphism (RFLP) analysis is the preferred method for VZV identification. Similar methods, such as long-distance PCR (33), single-strand conformation polymorphism analysis (22), fluorogenic PCR (TaqMan assay) (11), and other methods, can also distinguish between vaccine and wild-type strains of VZV. All these methods require postamplification processing (electrophoresis, enzyme-linked immunosorbent assay-based product detection), and many PCR methods cannot distinguish all Japanese wild-type strains from the Oka vaccine strain (10, 16, 22).
The single-step fluorescent resonance energy transfer (FRET) genotyping method described here uses rapid-cycle PCR coupled with resonance energy transfer with fluorophore-labeled hybridization probes (35, 36). The assay is fast and robust and differentiates the vaccine strain from wild-type strains with a high degree of specificity.
MATERIALS AND METHODS
Specimen collection and DNA purification.
VZV isolates (except those collected by the authors) were provided by John Zaia (City of Hope Hospital), Barbara Watson (Philadelphia Department of Public Health), Ann Arvin (Stanford University), Dominic Dwyer (Westmead Hospital), and Yuan-Xiang Meng, John Stewart, and Joe Esposito (Centers for Disease Control and Prevention). Material from 79 specimens was available for testing. Isolates originated from various geographic locations, including Japan (25 specimens), the United States (26 specimens), Australia (9 specimens), Democratic Republic of Congo (5 specimens), Chad (5 specimens), Nepal (5 specimens), China (3 specimens), and France (1 specimen), and were collected between 1976 and 1999. VZV DNAs from cells infected with the Oka vaccine strain (VARIVAX; Merck & Co., Inc., Rahway, N.J.) and three laboratory VZV strains (strains Webster, VZV11, and ROD) were also examined. Fifty of the DNA preparations were purified from virus isolates propagated in tissue culture. The remaining 29 clinical specimens were provided by general practitioners and infectious disease physicians and consisted of vesicular fluid air dried onto glass slides or cotton swabs or from scabs crusted over lesions that contained primary isolates of nonviable VZV. Genomic DNA was isolated from clinical samples with NucleoSpin Tissue Kits (CLONTECH Laboratories Inc., Palo Alto, Calif.). Purification of DNA from lysates of VZV-infected cells was performed as described previously (29, 33, 37). The DNA was resuspended in distilled water or 10 mM Tris (pH 8.0) per liter. DNA that was not used immediately was stored at 2 to 8°C for not longer than 1 week or was frozen at −20°C. To evaluate the FRET method, DNAs from VZV specimens that had been genotyped as wild-type strains by RFLP analysis of open reading frame (ORF) 62 (3, 21) amplicons were analyzed by the new method.
Design of primers and fluorogenic probes.
Primer PKVL2L (5′-GTG TCC GCT TTG AAC GCC CG-3′; positions 106409 to 106390, which are the corresponding positions of reference Dumas strain genome; GenBank accession number X04370) and primer PKVL7U (5′-AAC TCG CTG GCC CAA AGG TG-3′; positions 106109 to 106128) were used to amplify a 301-bp fragment of VZV ORF 62, which includes the polymorphic site at position 106262 (replacement of T with C in the Oka vaccine strain). The amplification primers were synthesized by standard phosphoramidite chemistry. The detection probe was an 18-mer oligonucleotide, labeled at the 5′ end with LightCycler Red 705 and modified at the 3′ end by phosphorylation to prevent extension. The sequence 5′-AGG TGG CCC AGG GAT GGA-3′ was complementary to the antisense strand of VZV ORF 62, with the polymorphic nucleotide 9 bases from the 3′ end (see Fig. 1). The 3′-fluorescein-labeled anchor probe was a 27-mer that binds at a distance of 4 bases 5′ to the detection probe (5′-GTT GCT GGT GTT GGA CGC GGT GGC CCT-3′). Both fluorophore-labeled probes were synthesized on an ABI model 394 DNA synthesizer (PE Biosystems, Foster City, Calif.) and were purified by reverse-phase high-pressure liquid chromatography. The amplification primers were described previously (21), and the base sequence for the Oka parental strain was published by Argaw et al. (3).
FIG. 1.
Relative orientations of the fluorophore-labeled anchor, the detection probe, and the PCR primer. The detection probe (De) spanning the polymorphic nucleotide at position 106262 of VZV ORF 62 was labeled at the 5′ end with LightCycler Red 705 and was phosphorylated at its 3′ end to block extension. The anchor probe (An) was labeled with fluorescein at its 3′ end. During annealing, both probes hybridize to the complementary sequence of the antisense strand. The proximity of the LightCycler Red 705 and fluorescein labels results in FRET, which is monitored at the end of each annealing step during PCR and continuously throughout recording of the melting curve (28). The ORF 62 polymorphism is a result of a T-to-C substitution at nucleotide 106262 in the Oka vaccine strain. This polymorphism creates an A-C mismatch between the antisense strand and the detection probe in the case of the Oka vaccine strain. This mismatch destabilizes the hybrid, which results in a decrease in the probe's melting temperature. In contrast, complete matching of the detection probe and the antisense strand results in a higher melting temperature of the hybrid for wild-type strains.
Amplification and mutation detection protocol.
PCR was performed by rapid cycling in a reaction volume of 20 μl with each primer at a concentration of 0.25 μM, detection probe at a concentration of 0.2 μM, 0.4 μM anchor probe, and 50 ng of genomic DNA. The LightCycler DNA Master Hybridization Probe buffer was used as a reaction buffer (Roche Molecular Biochemicals, Mannheim, Germany). This buffer was provided as a 10× stock solution containing nucleotides, Taq DNA polymerase, and 10 mM Mg2+. The final Mg2+ concentration in the reaction mixture was adjusted to 3 mM. The samples were loaded into glass capillary cuvettes (Roche Molecular Biochemicals, Mannheim, Germany) and were centrifuged to place the sample at the capillary tip before capping. To ensure a hot start for PCR, 0.32 μl of anti-Taq polymerase antibodies (Clontech Laboratories, Inc., Palo Alto, Calif.) was added to each reaction mixture. After an initial denaturation step and antibody inactivation at 94°C for 2 min, amplification was performed by using 40 cycles of denaturation (95°C for <1 s), annealing (57°C for 7 s), and extension (72°C for 15 s) on a LightCycler fluorometric thermal cycler (Roche Molecular Biochemicals, Mannheim, Germany). Fluorescence was measured at the end of the annealing period of each cycle to monitor the concentration of amplicon. After amplification was complete, a final melting curve was recorded by heating to 95°C for 2 min and then cooling to 50°C, followed by a 35-s hold before heating slowly at intervals of 0.2°C until a temperature of 80°C was attained. Fluorescence was measured continuously during the slow temperature rise to monitor the dissociation of the LightCycler Red 705-labeled detection probe. The fluorescence signal (F) was plotted in real time against the temperature (T) to produce melting curves for each sample (F versus T). Melting curves were then converted to melting peaks by plotting the negative derivative of F with respect to T against T (−dF/dT against T). The entire process required approximately 60 min (23).
Genotyping by RFLP analysis.
For confirmation of VZV wild-type genotypes, RFLP analysis was performed as described elsewhere (21). A 268-bp fragment encompassing a mutation in the Oka vaccine VZV strain was amplified from genomic DNA by using the primers described below. For fragment amplification, each oligonucelotide primer at a concentration of 0.1 μM (upper primer, PKVL_6U [5′-TTC CCA CCG CGG CAC AAA CA-3′], VZV genome position 106036; lower primer, PKVL_1L [5′-GGT TGC TGG TGT TGG ACG CG-3′], VZV genome position 106284) was used in a 100-μl reaction mixture containing PCR Gold buffer (50 mM KCl, 15 mM Tris-hydrochloride [pH 8.0]), 2.5 mM MgCl2, dATP, dCTP, dGTP, and dTTP each at a concentration of 200 μM, and 2.5 U of AmpliTaq Gold DNA polymerase (PE Biosystems and Roche Molecular Biochemicals, Indianapolis, Ind.). For amplification, 500 ng of a total DNA preparation from VZV-infected HLF cells was used as a template. For clinical specimens, PCR assays used a 1:100 portion of the DNA purified from a single lesion (scab or swab). An initial 15-min PCR hot-start step of 96°C was followed by 30 cycles of amplification (1 min at 94°C, 1 min at 72°C) and a final extension step at 72°C for 3 min. Reactions were performed in a Mastercycler gradient thermocycler (Eppendorf Scientific Inc., Westbury, N.Y.). The amplicons were resolved by electrophoresis in precast 4 to 20% gradient polyacrylamide gels in Tris-borate-EDTA buffer (Novex, San Diego, Calif.) and were stained with ethidium bromide to visualize the DNA. Restriction reactions were performed with 5 to 10 μl of the PCR product adjusted in the recommended endonuclease buffer and 10 U of SmaI (New England Biolabs, Inc., Beverly, Mass.). Endonuclease-cleaved DNA products were separated by gel electrophoresis as described above. For DNA size reference markers, 50- and 100-bp DNA ladders (Life Technologies Inc., GIBCO BRL, Gaithersburg, Md.) were used.
RESULTS
Detection and differentiation of vaccine from wild-type strains of VZV.
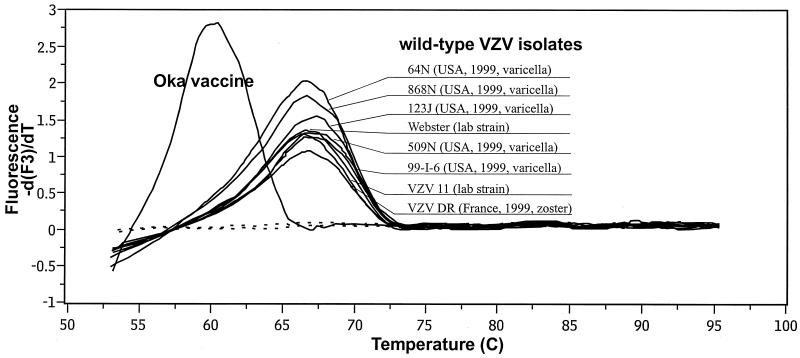
To assess the ability to detect and distinguish the ORF 62 sequence heterogeneity between the Oka vaccine strain and wild-type strains of VZV, 40 cycles of amplification were performed with genomic DNAs from two laboratory strains of VZV (strains Webster and VZV11), six clinical specimens (specimens 64N, 868N, 123J, 509N, 99-I-6, and VZV DR), and the Oka vaccine strain with the FRET detection system and the sequences described in Fig. 1. A template-free control specimen was also tested. Resonance energy transfer occurs during the annealing phase of each cycle as the fluorophore-labeled probes hybridize to the antisense strand of the amplicon. The process of hybridization and melting of the detection probe to the target coincides with the acquisition and the loss of the fluorescence signal, since the donor and acceptor probes are in close proximity only when the probes have annealed to the amplicon DNA. The fluorescence signals increased with each cycle in direct proportion to the accumulation of specific PCR product and generally rose above the background levels after 10 to 20 cycles (data not shown). By plotting the negative derivative of the fluorescence signal with temperature versus temperature (−dF/dT versus T), peaks were obtained at the respective melting temperatures (Fig. 2). Heteroduplex annealing of ORF 62 DNA from the Oka vaccine strain with the ORF 62 detection probe occurred at between 59 and 60°C. In contrast, annealing of the detection probe with DNA from any of the wild-type and laboratory strains took place at between 65 and 67°C. As such, the differences in ORF 62 probe annealing temperatures were more than sufficient to permit unequivocal differentiation of the Oka vaccine strain and other strains of VZV.
FIG. 2.
Genotyping of VZV with a fluorescent probe by derivative melting curve plot. Following amplification, a melting curve analysis was immediately performed. Data for the plot were obtained during the melting transition of the LightCycler Red 705-labeled detection probe from the amplified fragment.
No increase in fluorescence signal was observed in the absence of template. The assay is also VZV specific, since no signal developed with tissue culture material containing DNA from any of the following human herpesviruses: Epstein-Barr virus, cytomegalovirus, herpes simplex virus types 1 and 2, human herpesvirus 6 variants a and b, and human herpesvirus 8 (data not shown).
Comparison of FRET-based strain discrimination with RFLP methods.
To evaluate the practicability and reliability of the fluorescence genotyping in a clinical routine setting, 79 clinical and tissue culture-grown VZV samples were genotyped by the SmaI RFLP method and by the FRET-based method (Table 1). The PCR products and fragments obtained by RFLP analysis were of the expected sizes (data not shown). The specimens collected for this study represent circulating VZV strains from five of the six inhabited continents. Positive (wild-type and vaccine strain) DNA and negative controls were included in each test run. The genotypes determined by both methods were in complete concordance. The virus in 1 sample, which is thought to come from a patient with a vaccine-related case of varicella, was typed as the Oka vaccine strain, while the viruses in 78 samples were determined to be wild-type strains. For all 79 wild-type or laboratory strains, the detection probe annealed to the DNA template at between 65 and 67°C, indicating that the region complementary to the probe is exceedingly stable, apart from the single point mutation that defines the Oka vaccine strain.
TABLE 1.
Comparison of FRET-based PCR with conventional RFLP analysis
| Origin of isolate | Methoda | Genotype
|
|
|---|---|---|---|
| Wild-type VZV | Oka vaccine strain VZV | ||
| United States | FRET | 25 | 1 |
| RFLP | 25 | 1 | |
| Japan | FRET | 25 | 0 |
| RFLP | 25 | 0 | |
| Australia | FRET | 9 | 0 |
| RFLP | 9 | 0 | |
| Chad | FRET | 5 | 0 |
| RFLP | 5 | 0 | |
| Nepal | FRET | 5 | 0 |
| RFLP | 5 | 0 | |
| Congo | FRET | 5 | 0 |
| RFLP | 5 | 0 | |
| China | FRET | 3 | 0 |
| RFLP | 3 | 0 | |
| France | FRET | 1 | 0 |
| RFLP | 1 | 0 | |
FRET, FRET-based PCR with the ORF 62-based probes and primers described in Fig. 1; RFLP, SmaI digestion of the ORF 62 amplicon by RFLP analysis, as described in Materials and Methods.
Serial dilution of DNA specimens revealed that FRET-based melting curve analysis was more sensitive than conventional RFLP analysis (with UV transillumination) by as much as 40-fold (data not shown).
DISCUSSION
Amplification of targeted regions of the VZV genome followed by specific restriction endonuclease digestion (RFLP analysis) has become the most reliable and effective means for discrimination of the Oka vaccine strain and wild-type strains, an ability essential to the characterization of breakthrough infections among VZV vaccinees (5, 34). Nonetheless, the time required to complete such assays (usually, 2 full working days) has limited their usefulness in the reporting of test results for clinical specimens. In the present study we used probe hybridization and FRET to monitor specific product accumulation during rapid-cycle DNA amplification as a means for discrimination of vaccine and wild-type VZV strains. In contrast to conventional RFLP analysis, these assays can be completed in 5 h or less, depending on the time required to prepare the DNA from a specimen.
In this study we looked at only a single specimen of the Oka vaccine strain obtained from a lot of commercial vaccine, raising the possible concern that the ORF 62 mutation at nucleotide 106262 is not consistently maintained in the vaccine. However, others have examined VZV from a variety of Oka vaccine lots and consistently detected this mutation (3; N. Kraiouchkine, personal communication). In addition, the virus in one U.S. specimen, obtained from a vesicular lesion on a child who developed varicella several weeks postvaccination, was identified as a vaccine strain by the two methods used in the present study as well as by the RFLP method described by LaRussa et al. (16).
Several PCR-based methods have been used to genotype VZV strains (5, 7, 28, 32, 34), but each of the methods has some drawbacks. A limited number of point mutations in the vaccine strain or wild-type strains of VZV that can be detected with specific restriction endonucleases have been identified, and primers that amplify these regions have been described (11, 16, 29, 33). Some of these are more effective than others at discriminating vaccine strain from wild-type virus (16); among the most useful are mutations identified in ORFs 38 and 54 and, more recently, in ORF 62 (3, 16, 21). Preferentional homoduplex formation is time-consuming, and reliable results are crucially dependent on the quality of the PCR products and the hybridization conditions (25). The single-strand conformational polymorphism method has also been used by some laboratories for genotyping, but this method is laborious and yields results that are sometimes difficult to interpret. Most of the protocols currently in use require additional steps for product detection and identification, increasing the time and expense of the assay. In addition to the inherent risk of false-positive amplification, the performance of the sequence-specific primers in these assays must be assured by amplifying internal control fragments. Techniques based on RFLP analysis need PCR conditions of sufficient specificity to produce a clean amplification product that can be enzymatically digested and unambiguously analyzed by electrophoresis. Furthermore, the interpretation of results by this approach may be complicated by incomplete digestion of amplimers. The ligase chain reaction technique (14), while combining high degrees of sensitivity and specificity with the potential of automation, still requires several postamplification procedures, including a ligation reaction and in some protocols subsequent enzyme-linked immunosorbent assay-based detection.
The FRET-based PCR method described here is performed in a single reaction vessel, with no further manipulation of the amplified product required. The sequence heterogeneity is detected directly, in real time, by using a short DNA probe that overlies the point mutation. Since the reaction is carried out in a single step in a closed system, there is no risk of carryover contamination following PCR amplification. As such, by conducting the DNA extraction step and preparing PCR master mixtures in separate locations and by using conventional precautions, such as wearing disposable gloves and gowns, the risk of sample DNA contamination can be virtually eliminated in this procedure.
The FRET-based PCR equipment used for this study uses capillary reaction vessels, with adjustment of the cycling temperatures done by alternating heated and ambient air. The result is that cycling times are substantially faster than those achievable with a block or water-based thermocycler. Because of the high surface area-to-volume ratio of the capillaries, combined with the rapid temperature shifts made possible by this method, a single PCR cycle can be accomplished in less than 1 min. A complete run of 40 cycles can be completed in 30 min. The fluorescent probes hybridize to the amplified product during the annealing phase, which allows real-time collection of the FRET signal. Sequence heterogeneity can be distinguished by melting point temperature analysis post-PCR amplification.
The detection of the VZV ORF 62 mutation at the nucleic acid level without restriction enzyme digestion represents a novel approach to VZV genotyping by PCR. The robustness and reliability of fluorescence genotyping was documented by the complete concordance of the genotypes determined by conventional ORF 62-based RFLP analysis and the new protocol for all 79 specimens tested. Amplification of viral DNA extracted directly from vesicular fluid or scabs eliminates the need for virus isolation and requires only a small quantity of material. The benefits of homogeneous detection systems have long been recognized, but such systems have not been commercially available. Recently, several fluorescence-based methods for the typing of biallelic systems have been described (20). Using the approach described here, we were able to genotype 32 VZV specimens in 60 min without restriction enzyme digestion or electrophoresis. This FRET-based method combines simple routine processing and rapid analysis and therefore affords both high-throughput genotyping and rapid results. Furthermore, as the hands-on time is shorter than that for any other technique used so far, this method results in VZV genotyping in an economic manner in laboratories equipped to perform FRET-based assays. The feasibility of specific PCR product detection without electrophoresis or restriction endonuclease digestion makes this method attractive for studies of the molecular epidemiology of VZV.
ACKNOWLEDGMENTS
The excellent technical assistance of Dominique Rollin is gratefully acknowledged. Ann Arvin, Dominic Dwyer, Joe Esposito, John Stewart, Barbara Watson, and John Zaia kindly provided most of the non-Japanese VZV isolates used in these studies. We also thank John O'Connor for editing the manuscript.
REFERENCES
- 1.American Academy of Pediatrics, Committee on Infectious Diseases. Recommendations for the use of live attenuated varicella vaccine. Pediatrics. 1995;95:791–796. [PubMed] [Google Scholar]
- 2.American Academy of Pediatrics, Committee on Infectious Diseases. Varicella vaccine update. Pediatrics. 2000;105:136–141. [PubMed] [Google Scholar]
- 3.Argaw T, Cohen J L, Klutch M, Lekstrom K, Yoshikawa T, Asano Y, Krause P R. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J Infect Dis. 2000;181:1153–1157. doi: 10.1086/315335. [DOI] [PubMed] [Google Scholar]
- 4.Arvin A M, Gershon A A. Live attenuated varicella vaccine. Annu Rev Microbiol. 1996;50:59–100. doi: 10.1146/annurev.micro.50.1.59. [DOI] [PubMed] [Google Scholar]
- 5.Asano Y, Suga S, Yoshikawa T, Kobayashi I, Yazaki T, Shibata M, Tsuzuki K, Ito S. Experience and reason: twenty-year follow-up of protective immunity of the Oka strain live varicella vaccine. Pediatrics. 1994;94:524–526. [PubMed] [Google Scholar]
- 6.Gershon A A. Varicella-zoster virus: prospects for control. Adv Pediatr Infect Dis. 1995;10:93–124. [PubMed] [Google Scholar]
- 7.Gershon A A, LaRussa P, Hardy I, Steinberg S, Silverstein S. Varicella vaccine: the American experience. J Infect Dis. 1992;166(Suppl. 1):S63–S68. doi: 10.1093/infdis/166.supplement_1.s63. [DOI] [PubMed] [Google Scholar]
- 8.Gershon A A, Steinberg S P, LaRussa P, Ferrara A, Hammerschlag M, Gelb L. Immunization of healthy adults with live attenuated varicella vaccine. J Infect Dis. 1988;158:132–137. doi: 10.1093/infdis/158.1.132. [DOI] [PubMed] [Google Scholar]
- 9.Hardy I B, Gershon A, Steinberg S, LaRussa P. The incidence of zoster after immunization with live attenuated varicella vaccine: a study in children with leukemia. N Engl J Med. 1991;325:1545–1550. doi: 10.1056/NEJM199111283252204. [DOI] [PubMed] [Google Scholar]
- 10.Hawrami K, Breuer J. Analysis of United Kingdom wild-type strains of varicella-zoster virus: differentiation from the Oka vaccine strain. J Med Virol. 1997;53:60–62. doi: 10.1002/(sici)1096-9071(199709)53:1<60::aid-jmv10>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Hawrami K, Breuer J. Development of a fluorogenic polymerase chain reaction assay (TaqMan) for the detection and quantitation of varicella zoster virus. J Virol Methods. 1999;79:33–40. doi: 10.1016/s0166-0934(98)00176-1. [DOI] [PubMed] [Google Scholar]
- 12.Hawrami K, Hart L J, Pereira F, Argent S, Bannister B, Bovill B, Carrington D, Ogilvie M, Rawstorne S, Tryhorn Y, Breuer J. Molecular epidemiology of varicella-zoster virus in East London, England, between 1971 and 1995. J Clin Microbiol. 1997;35:2807–2809. doi: 10.1128/jcm.35.11.2807-2809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland P, Isaacs D, Moxon E R. Fatal neonatal varicella infection. Lancet. 1986;ii:1156. doi: 10.1016/s0140-6736(86)90556-8. [DOI] [PubMed] [Google Scholar]
- 14.Kalin I, Shephard S, Candrian U. Evaluation of the ligase chain reaction (LCR) for the detection of point mutations. Mutat Res. 1992;283:119–123. doi: 10.1016/0165-7992(92)90143-6. [DOI] [PubMed] [Google Scholar]
- 15.Koskiniemi M, Mannonen L, Kallio A, Vaheri A. Luminometric microplate hybridization for detection of varicella-zoster virus PCR product from cerebrospinal fluid. J Virol Methods. 1997;63:71–79. doi: 10.1016/s0166-0934(96)02116-7. [DOI] [PubMed] [Google Scholar]
- 16.LaRussa P, Lungu O, Hardy I, Gershon A, Steinberg S P, Silverstein S. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J Virol. 1992;66:1016–1020. doi: 10.1128/jvi.66.2.1016-1020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaRussa P, Steinberg S, Arvin A, Dwyer D, Burgess M, Menegus M, Rekrut K, Yamanishi K, Gershon A. Polymerase chain reaction and restriction fragment length polymorphism analysis of varicella-zoster virus isolates from the United States and other parts of the world. J Infect Dis. 1998;178:S64–S66. doi: 10.1086/514267. [DOI] [PubMed] [Google Scholar]
- 18.LaRussa P, Steinberg S, Meurice F, Gershon A. Transmission of vaccine strain varicella-zoster virus from a healthy adult with vaccine-associated rash to susceptible household contacts. J Infect Dis. 1997;176:1072–1075. doi: 10.1086/516514. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence R, Gershon A A, Holzman R, Steinberg S P. The risk of zoster after varicella vaccination in children with leukemia. N Engl J Med. 1988;318:543–548. doi: 10.1056/NEJM198803033180904. [DOI] [PubMed] [Google Scholar]
- 20.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. Genome Res. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 21.Loparev V L, Argaw T, Krause P R, Takayama M, Schmid D S. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J Clin Microbiol. 2000;38:3156–3160. doi: 10.1128/jcm.38.9.3156-3160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori C, Takahara R, Toriyama T, Nagai T, Takahashi M, Yamanishi K. Identification of the Oka strain of the live attenuated varicella vaccine from other clinical isolates by molecular epidemiologic analysis. J Infect Dis. 1998;178:35–38. doi: 10.1086/515598. [DOI] [PubMed] [Google Scholar]
- 23.Morrison L E. Detection of energy transfer and fluorescence quenching. In: Kricka L J, editor. Nonisotopic DNA probe techniques. San Diego, Calif: Academic Press, Inc.; 1992. pp. 311–352. [Google Scholar]
- 24.Nikkels A F, Delvenne P, Sadzot-Delvaux C, Debrus S, Piette J, Rentier B, Lipcsei G, Quatresooz P, Pierard G E. Distribution of varicella-zoster virus and herpes simplex virus in disseminated fatal infections. J Clin Pathol. 1996;49:243–248. doi: 10.1136/jcp.49.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka T, Matsunaga H, Tokunaga K, Mitsunaga S, Juji T, Yamane A. A simple method for detecting single base substitutions and its application to HLA-DPB1 typing. Nucleic Acids Res. 1994;22:1541–1547. doi: 10.1093/nar/22.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozaki T, Kajita Y, Asano Y, Aono T, Yamanishi K. Detection of varicella-zoster virus DNA in blood of children with varicella. J Med Virol. 1994;44:263–265. doi: 10.1002/jmv.1890440309. [DOI] [PubMed] [Google Scholar]
- 27.Plotkin S A. Varicella vaccine. Pediatrics. 1996;97:251–253. [PubMed] [Google Scholar]
- 28.Shiraki K, Horiuchi K, Asano Y, Yamanishi K, Takahashi M. Differentiation of Oka varicella vaccine strain from wild varicella-zoster virus strains isolated from vaccinees and household contact. J Med Virol. 1991;33:128–132. doi: 10.1002/jmv.1890330212. [DOI] [PubMed] [Google Scholar]
- 29.Takada M, Suzutani T, Yoshida I, Matoba M, Azuma M. Identification of varicella-zoster virus strains by PCR analysis of three repeat elements and a PstI-site-less region. J Clin Microbiol. 1995;33:658–660. doi: 10.1128/jcm.33.3.658-660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi M. Clinical overview of varicella vaccine: development and early studies. Pediatrics. 1986;78:736–741. [PubMed] [Google Scholar]
- 31.Takahashi M, Otsuka T, Okuno Y, Asano Y, Yazaki T. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet. 1974;ii:1288–1290. doi: 10.1016/s0140-6736(74)90144-5. [DOI] [PubMed] [Google Scholar]
- 32.Takayama N, Minamitani M, Takayama M. High incidence of breakthrough varicella observed in healthy Japanese children immunized with live attenuated varicella vaccine (Oka strain) Acta Paediatr Jpn. 1997;39:663–668. doi: 10.1111/j.1442-200x.1997.tb03664.x. [DOI] [PubMed] [Google Scholar]
- 33.Takayama M, Takayama N, Inoue N, Kameoka Y. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J Clin Microbiol. 1996;34:2869–2874. doi: 10.1128/jcm.34.12.2869-2874.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson B M, Piercy S A, Plotkin S A, Starr S E. Modified chickenpox in children immunized with the Oka/Merck varicella vaccine. Pediatrics. 1993;91:17–22. [PubMed] [Google Scholar]
- 35.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 36.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The LightCycler. A microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida M, Tamura T, Hiruma M. Analysis of strain variation of R1 repeated structure in varicella-zoster virus DNA by polymerase chain reaction. J Med Virol. 1999;58:76–78. [PubMed] [Google Scholar]