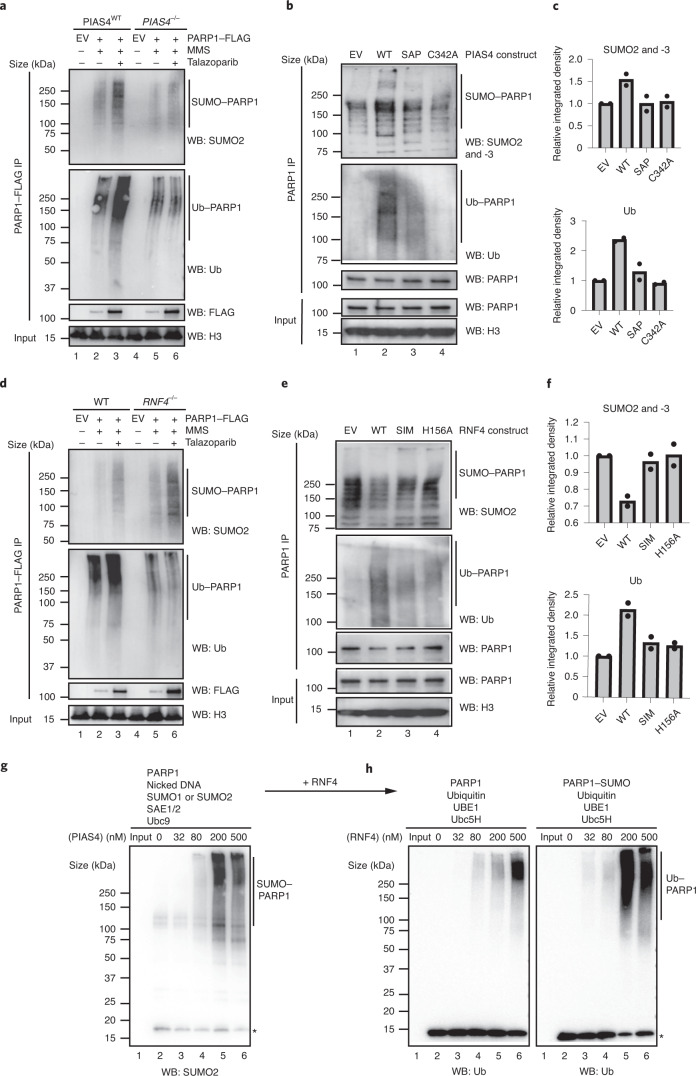
Fig. 3. Trapped PARP1 is modified in a PIAS4- and RNF4-dependent manner.
a, PARP1 is SUMOylated in a PIAS4-dependent manner in vivo. Wild-type and PIAS4–/– HCT116 cells were transfected with FLAG–PARP1-expressing plasmid, exposed to trapping and the chromatin-bound PARP1 was investigated for SUMOylation and ubiquitylation. The levels of SUMO1 (Extended Data Fig. 4a), SUMO2 and ubiquitin were reduced in the PIAS4–/– cells (see Extended Data Fig. 4b,c for the total ubiquitin input and quantification of the blots, respectively). b, PIAS4–/– HCT116 cells were transfected with different PIAS4-expressing constructs for 48 h, followed by 30 min talazoparib (10 µM) treatment in the presence of 0.01% MMS and PARP1 immunoprecipitation. c, Abundance of SUMO2 and -3 (top)- and ubiquitin (bottom)-modified PARP1 in b. b,c, Data represent two biological replicates. SAP, PIAS4 with deleted SAP domain; and C342A, catalytic dead PIAS4. d, Similarly to a, trapped PARP1 was purified from the chromatin fraction of wild-type and RNF4–/– MCF7 cells. The PARP1 ubiquitylation levels were reduced in the RNF4–/– cells, whereas SUMO1- (Extended Data Fig. 4e) and SUMO2-ylation were increased (see Extended Data Fig. 4f,g for the total ubiquitin input and quantification of the blots). e, RNF4–/– MCF7 cells were transfected with different RNF4-expressing plasmids for 48 h and processed as in b. f, Abundance of SUMO2 and -3 (top)- and ubiquitin (bottom)-modified PARP1 in e. e,f, Data represent two biological replicates. SIM, RNF4 with deleted SUMO-interacting motif; and H156A, catalytic dead RNF4. g, PIAS4 mediates PARP1 SUMOylation in vitro. Recombinant PARP1 was incubated with nicked DNA, SUMO1 or SUMO2, SAE1 and -2, Ubc9 and an increasing concentration of PIAS4. PIAS4 led to a concentration-dependent increase of SUMOylation (Extended Data Fig. 5c). *Free SUMO2. h, RNF4 mediates PARP1 ubiquitylation in a SUMO-dependent manner in vitro. PARP1 SUMOylation reactions were supplemented with ubiquitin, UBE1, Ubc5H and an increasing concentration of RNF4. SUMOylated PARP1 was a better substrate for ubiquitylation. *Free ubiquitin. a,d,g,h, Data shown represent two independent experiments with similar results. EV, empty vector; IP, immunoprecipitation; WB, western blot; and WT, wild type.