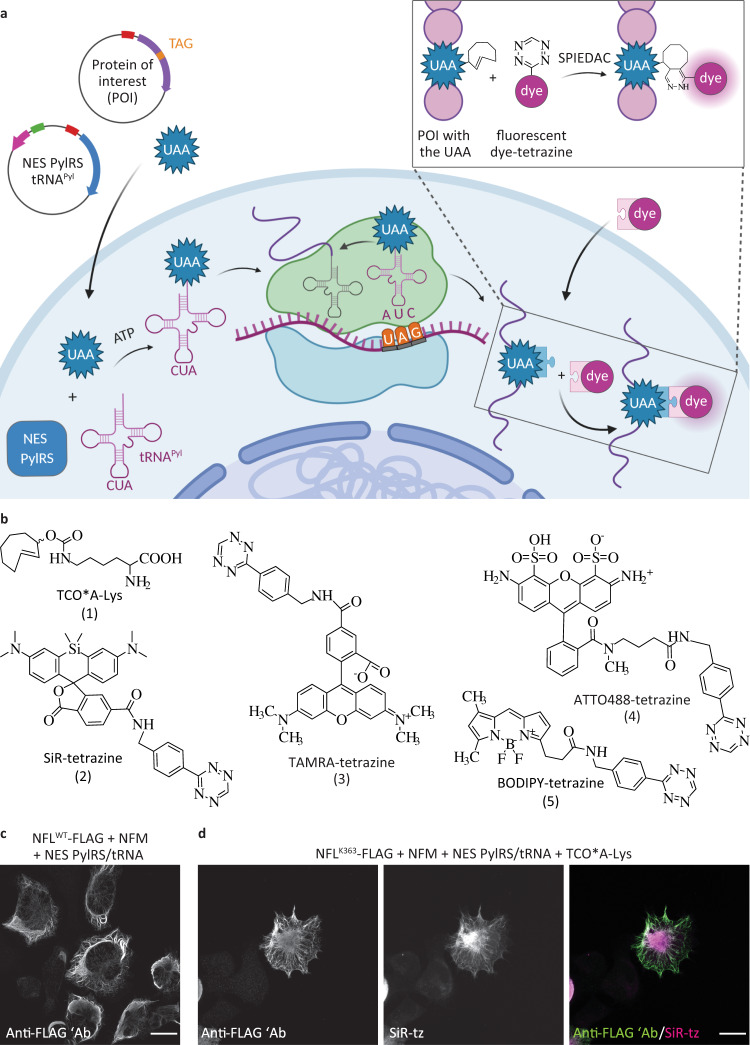
Fig. 1. Genetic code expansion and click labeling of neurofilament light chain (NFL) in ND7/23 neuroblastoma cells.
a A schematic representation of genetic code expansion and labeling of proteins by click chemistry. Cells are transfected with plasmids bearing genes that encode for NES PylRS (a pyrrolysyl-tRNA synthetase with a nuclear export signal) and tRNACUAPyl, as well as a TAG stop codon-containing gene that encodes for the protein of interest (POI). The NES PylRS charges tRNACUAPyl with the unnatural amino acid (UAA), this charged tRNACUAPyl recognizes the UAG stop codon in the mRNA encoding the POI, and UAA is co-translationally incorporated into the POI. In the subsequent step, a tetrazine derivative of a fluorescent dye reacts with the UAA by strain-promoted inverse electron-demand Diels–Alder cycloaddition (SPIEDAC) reaction, and the dye is attached directly to the POI. b Chemical structures of UAAs and tetrazine dyes that were used to obtain the data shown in the main figures. c ND7/23 cells expressing neurofilament medium-chain (NFM), NES PylRS/tRNACUAPyl, and wild-type NFLWT-FLAG, stained with an anti-FLAG antibody, followed by Alexa Fluor (AF) 488-conjugated secondary antibody. d ND7/23 cells expressing NFLK363TAG-FLAG, NFM, and NES PylRS/tRNACUAPyl. Cells were incubated overnight with TCO*A-Lys and then click labeled with silicon rhodamine-tetrazine (SiR-tz). Afterward, cells were fixed and stained with the anti-FLAG antibody, followed by AF488-conjugated secondary antibody. Z-stack images were acquired with a confocal scanning microscope, and are shown as maximum intensity projections. Scale bars: 20 µm (c, d). Data were collected from three independent experiments (c, d). Scheme in panel a was partially created with BioRender.com.