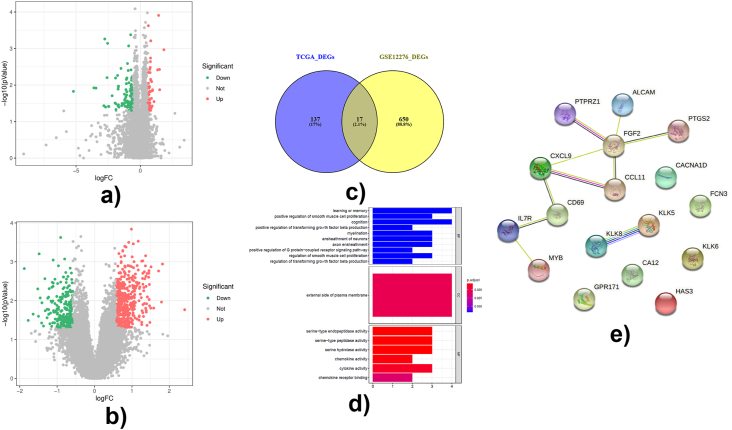
Fig. 1.
Identification and functional analysis of differentially expressed metastatic genes. Differentially expressed genes were identified by limma R package using |fold change| > 1.5 and P-value < 0.05 as criteria. Functional analysis was performed by clusterProfiler R package.(A) Volcano plot of differentially expressed genes between 907 M0 and 22 M1 breast cancer samples. The red dots were upregulated DEGs, the green dots were downregulated DEGs, and the grey dots were genes without significant difference. (B)Volcano plot of differentially expressed genes between 16 breast cancer patients with brain metastases and 188 breast cancer patients with other metastases. The red dots were upregulated DEGs, the green dots were downregulated DEGs, and the grey dots were genes without significant difference. (C) Venn plot showed the intersection of 17 differentially expressed metastatic genes in breast cancer. (D) Top 10 GO terms of differentially expressed metastatic genes, including biological process (BP), cellular component (CC) and molecular function (MF) were identified by clusterProfiler R package and were shown in the bar chart. (E) The protein-protein interaction network of DEMGS analyzed by STRING database. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)