Abstract
DNA N6-adenine methylation (6mA), as a novel adenine modification existing in eukaryotes, shows essential functions in embryogenesis and mitochondrial transcriptions. ALKBH1 is a demethylase of 6mA and plays critical roles in osteogenesis, tumorigenesis, and adaptation to stress. However, the integrated biological functions of ALKBH1 still require further exploration. Here, we demonstrate that knockdown of ALKBH1 inhibits adipogenic differentiation in both human mesenchymal stem cells (hMSCs) and 3T3-L1 preadipocytes, while overexpression of ALKBH1 leads to increased adipogenesis. Using a combination of RNA-seq and N6-mA-DNA-IP-seq analyses, we identify hypoxia-inducible factor-1 (HIF-1) signaling as a crucial downstream target of ALKBH1 activity. Depletion of ALKBH1 leads to hypermethylation of both HIF-1α and its downstream target GYS1. Simultaneous overexpression of HIF-1α and GYS1 restores the adipogenic commitment of ALKBH1-deficient cells. Taken together, our data indicate that ALKBH1 is indispensable for adipogenic differentiation, revealing a novel epigenetic mechanism that regulates adipogenesis.
Keywords: ALKBH1, DNA N6-adenine demethylase, mesenchymal stem cells, adipogenesis, HIF-1
Abbreviations: 6mA, N6-adenine methylation; CCK8, cell counting kit-8; BSA, bovine serum albumin; GYS1, glycogen synthase 1; HIF-1, hypoxia-inducible factor-1; hMSCs, human mesenchymal stem cells; iWAT, inguinal white adipose tissue; MSC, mesenchymal stem cell; SASP, senescence-associated secretory phenotype; siRNA, small interfering RNAs
Mesenchymal stem cells (MSCs) are capable of self-renewal and pluripotent differentiation, which eventually develop into specific types of somatic cells. The appropriate balance on cell fate decisions requires precise regulation of transcriptional and epigenetic networks (1, 2). Under the pathological conditions, an increasing number of cells undergo a shift from osteogenesis to adipogenesis (3, 4), resulting in the accumulation of lipid droplets and various metabolic disorders (5, 6). These changes lead to tissue inflammation, senescence-associated secretory phenotype (SASP), and osteoporosis (5, 7, 8, 9). Extensive efforts have been devoted to elucidating the pluripotency MSCs maintained during differentiation. Although plenty of literatures are trying to determine these phenotypes in various mechanisms, the connection between DNA methylations and pluripotency MSCs remains unclear.
Epigenetic regulations mainly refer to DNA methylation, histone modifications, noncoding RNAs, transcription factor binding, and chromatin remodeling (10, 11). These modifications influenced by various stimuli are of paramount importance on cell lineage decisions (12, 13, 14). Abnormal methylation leads to tumorigenesis, neuronal development retardation, and metabolic disorders (15, 16, 17). Of note, human adipogenesis and osteogenesis are also relevant with DNA methylation alterations across all genomic segments (18, 19). In recent years, DNA N6-adenine methylation (6mA), enzyme-mediated by N6AMT1 and ALBKH1, has been discovered to affect mammalian development (20, 21).
ALKBH1, a 2-oxoglutarate and Fe2+-dependent hydroxylase, has been defined as a DNA demethylase (20). Studies show that ALKBH1 mediates the demethylation of mRNA, tRNA, and methylated lysine of histone H2A (22, 23, 24). Methylated nucleotides of DNA are the main substrates of ALKBH1 (20, 24). Aberrant 6mA level upon Alkbh1 knockout mice leads to prolonged-expression of pluripotency markers, retardation of embryogenesis, and differentiation of MSCs, thus causing neurogenesis disorders, sex-ratio distortion, infertility, and multiple defects in the eyes, skeleton, and genitals (22, 25, 26). Our previous study showed that ALKBH1 enhanced osteogenic differentiation by demethylating 6mA at the promoter area of ATF4 (19). To elucidate the network of 6mA in human diseases, we explore the relationship between ALKBH1 and adipose homeostasis, which is yet unclear.
In this study, we show that ALKBH1 promotes adipogenic differentiation of hMSCs and 3T3-L1 cells, leading to excessive accumulation of adipose in mice. Mechanically, ALKBH1 demethylates 6mA of HIF-1α and GYS1 to augment HIF-1 pathway activation. Our findings provide a new insight into the pivotal regulatory role of 6mA in differentiation and diseases.
Results
ALKBH1 is upregulated during adipogenic differentiation
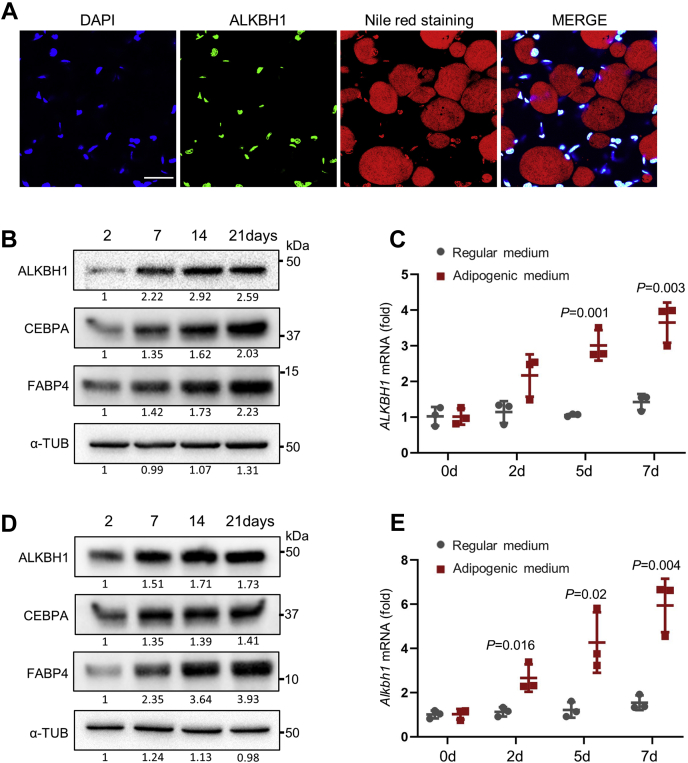
Firstly, we performed immunofluorescent staining of ALKBH1 and Nile red staining at the inguinal white adipose tissue (iWAT) of C57BL/6 mice and found that ALKBH1 was highly expressed in the nuclei of mature adipocytes (Fig. 1A). In addition, we induced hMSCs and 3T3-L1 preadipocytes toward mature adipocytes and observed increased protein levels of ALKBH1, as well as adipogenic essential proteins CEBPA and FABP4 (Fig. 1, B and D), The mRNA of ALKBH1 in both cells started to elevate from the early time of induction (Fig. 1, C and E).
Figure 1.
ALKBH1 is upregulated during adipogenic differentiation.A, immunofluorescence staining of ALKBH1 and Nile red staining in the iWAT of C57BL/6J mice. ALKBH1 is expressed widely in adipose. Scale bar: 30 μm. B, Western blot analysis of ALKBH1 and adipogenic markers in hMSCs cells during adipogenic differentiation. C, qRT-PCR analysis of mRNA level of ALKBH1 in hMSCs after adipogenic induction (n = 3). D, Western blot analysis of ALKBH1, CEBPA, and FABP4 in 3T3-L1 cells during adipogenic differentiation. E, qRT-PCR analysis of the expression of Alkbh1 in 3T3-L1 preadipocytes during adipogenic differentiation (n = 3). The p values were calculated by two-tailed Student's t test. Scatter plots show individual data points ± SD.
Depletion of ALKBH1 inhibits adipogenic differentiation in vitro
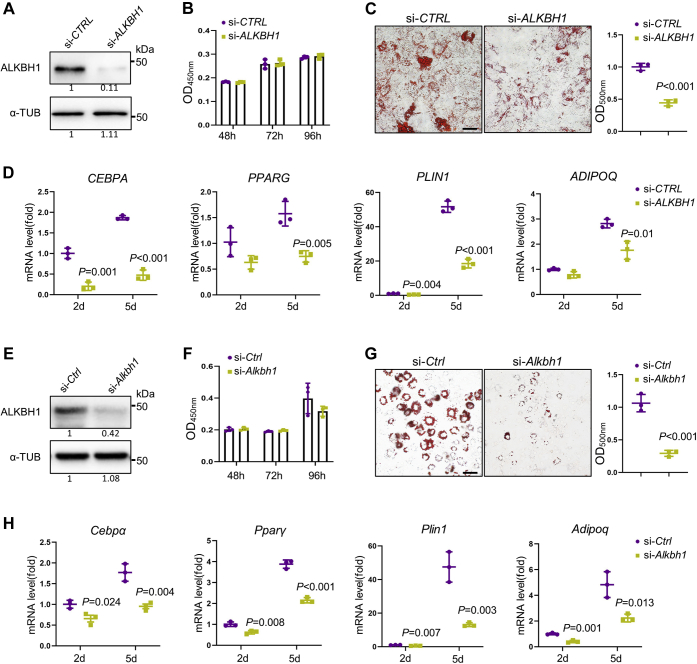
Next, we depleted ALKBH1 in hMSCs using small interfering RNAs (siRNA) and confirmed the knockdown efficiency by Western blot (Fig. 2A). The cell viability was not significantly affected after ALKBH1 depletion as checked by cell counting Kit-8 (CCK8) assay (Fig. 2B). After induction for 2 weeks, the incompetent adipogenic differentiation ability of ALKBH1-deficient hMSCs was evidenced by weaker Oil red O staining and less mature adipocytes (Fig. 2C), as well as the downregulated expression of adipogenic-related genes CEBPA, PPARG, PLIN1, and ADIPOQ (Fig. 2D).
Figure 2.
Depletion of ALKBH1 inhibits adipogenic differentiation in vitro.A, Western blot analysis confirms successful knockdown of ALKBH1 in hMSCs (n = 3). B, CCK8 assay shows the viability of hMSCs is not affected after ALKBH1 depletion (n = 3). The absorbance was analyzed at 450 nm at 48, 72, and 96 h. C, representative images and quantitative analysis of oil red O staining. hMSCs were transfected with siRNA and subjected to adipogenic induction for 14 days. Lipid droplets were dissolved by isopropanol and quantified at 500 nm (n = 3). Scale bar: 25 μm. D, qRT-PCR analysis of the mRNA expression of adipogenic related genes CEBPA, PPARG, PLIN1, ADIPOQ after 2- and 5-day adipogenic differentiation (n = 3). E, Western blot shows successful knockdown of ALKBH1 in 3T3-L1 cells. F, CCK8 assay of viability of 3T3-L1 cells after knockdown of Alkbh1. G, representative images and quantitation of oil red O staining in 3T3-L1 cells after induction of 14 days (n = 3). Scale bar: 25 μm. H, qRT-PCR analysis of Cebpα, Pparγ, Plin1, Adipoq in Alkbh1-knockdown 3T3-L1 cells (n = 3). The p values were calculated by two-tailed Student's t test. Scatter plots show individual data points ± SD.
Likewise, we knocked down Alkbh1 in 3T3-L1 cells (Fig. 2E) and found that it barely affected the cell proliferation (Fig. 2F), but inhibited the adipogenic differentiation of 3T3-L1 cells. The decreased differential potential was also demonstrated by Oil Red O staining (Fig. 2G) and qPCR analysis of the expression of adipogenic genes Cebpα, Pparγ, Plin1 and Adipoq (Fig. 2H).
Overexpression of ALKBH1 promotes adipogenesis
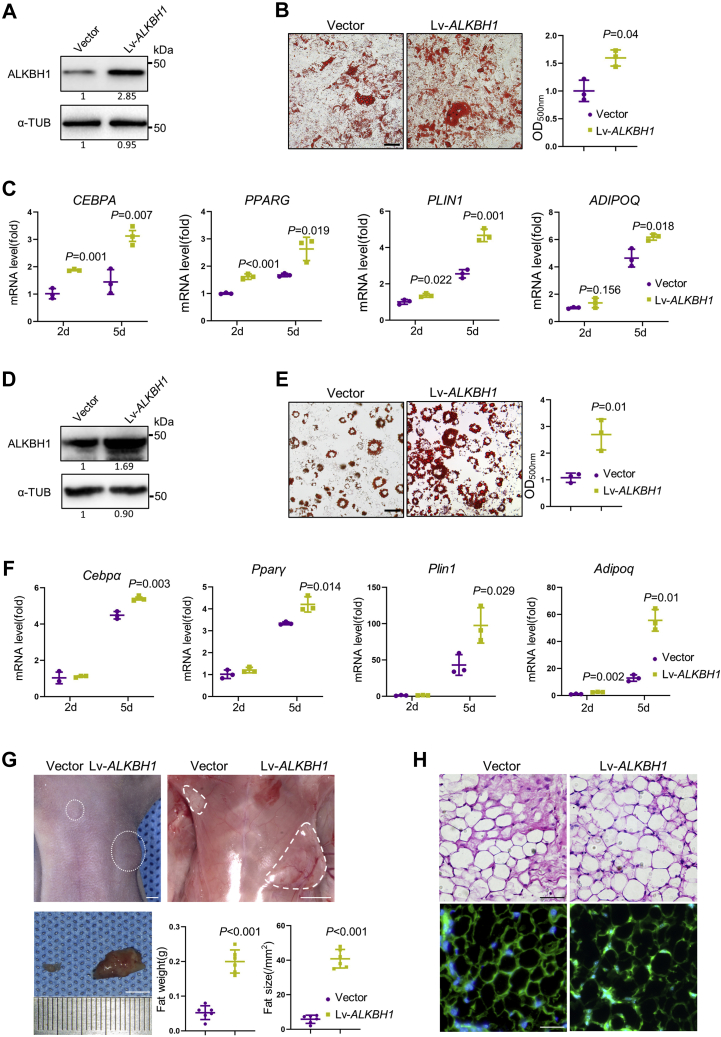
Next, we successfully infected hMSCs with control (Vector) or ALKBH1-overexpressing (Lv-ALKBH1) lentivirus (Fig. 3A). Oil red O staining revealed that Lipid accumulation is markedly increased in Lv-ALKBH1 group compared with control group (Fig. 3B). In addition, the mRNA levels of adipogenic related genes, such as CEBPA, PPARG, PLIN1, and ADIPOQ, were significantly elevated in the Lv-ALKBH1 group after 2 and 5 days of induction (Fig. 3C). Besides, we overexpressed ALKBH1 in 3T3-L1 cells (Fig. 3D) and observed increased intensity of oil red O staining (Fig. 3E) and elevated expression of adipocyte-specific genes, including Cebpα, Pparγ, Plin1, and Adipoq (Fig. 3F).
Figure 3.
Overexpression of ALKBH1 promotes adipogenesis.A, Western blot confirms the successful overexpression of ALKBH1 in hMSCs. B, quantitative analysis of oil red O staining in hMSCs after 14 days of adipogenic differentiation (n = 3). Scale bar: 25 μm. C, qRT-PCR analysis of adipogenic related genes CEBPA, PPARG, PLIN1, ADIPOQ in ALKBH1-overexpressed hMSCs (n = 3). D, Western blot shows overexpression of ALKBH1 in 3T3-L1 cells. E, representative images and the OD values of oil red O staining in 3T3-L1 cells (n = 3). Scale bar: 25 μm. F, qRT-PCR analysis of Cebpα, Pparγ, Plin1, Adipoq in ALKBH1-overexpressed 3T3-L1 cells (n = 3). G, macroscopic identification and quantitative analysis of ectopic fats after 6 weeks of sternal subcutaneous injection. Nude mice were bilateral transplanted with Vector and Lv-ALKBH1 3T3-L1 cells (n = 6). Scale bar: 5 mm. H, hematoxylin and eosin staining (top) and GFP staining (bottom) of fat pads isolated from nude mice. Scale bar: 25 μm. The p values were calculated by two-tailed Student's t test. Scatter plots show individual data points ± SD.
In addition, we performed an ectopic fat formation test by transplanting control and ALKBH1-overexpressing 3T3-L1 cells into bilateral sternum of nude mice (1). General observations showed that the fat pad at the Lv-ALKBH1 side is larger and weighs four times as much as the other side (Fig. 3G). HE staining and GFP immunofluorescence confirmed the source of adipose and showed the increasing capacity of ALKBH1-overexpressing 3T3-L1 cells (Fig. 3H).
ALKBH1 regulates HIF-1 pathway
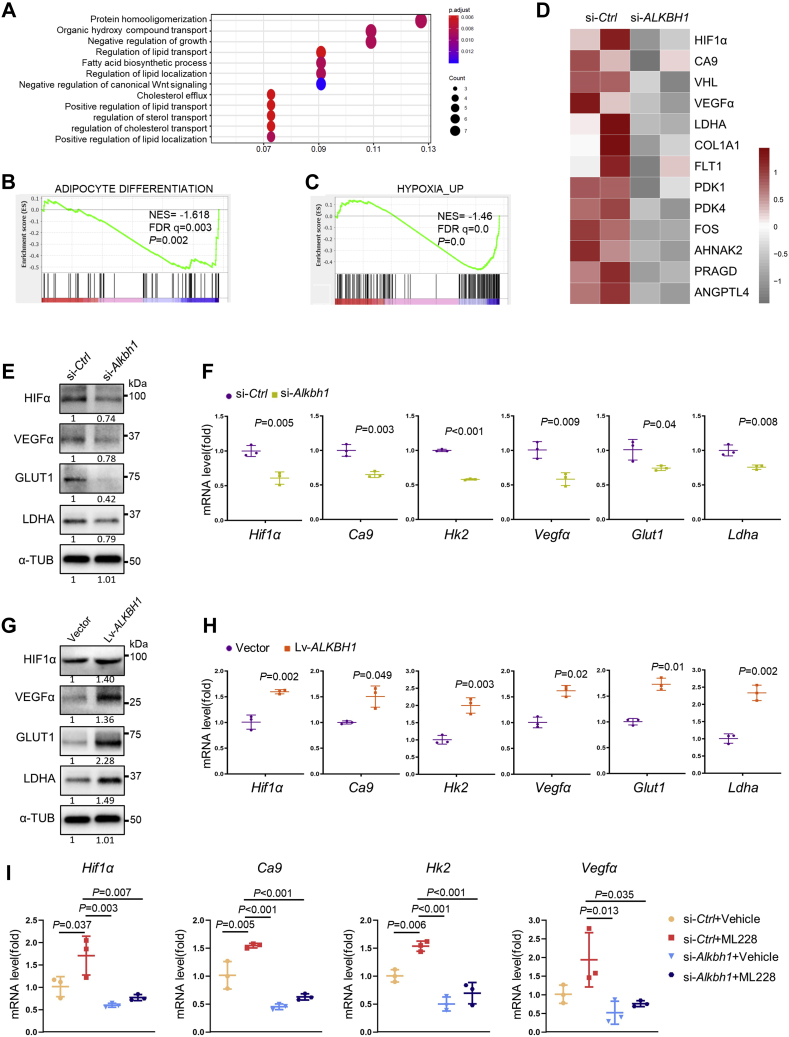
To elucidate the underlying mechanism, we performed RNA sequencing analysis on ALKBH1 knockdown hMSCs versus the control cells after 3-day adipogenic induction. We screened 330 downregulated genes and 166 upregulated genes in total. The Gene Ontology (GO) analysis interprets enriched biological functions of ALKBH1. ALKBH1 knockdown greatly downregulated the synthesis and transport of lipid (Fig. 4A). Gene set enrichment analysis (GSEA) confirmed the disruption of adipogenesis in ALKBH1-deficient hMSCs (Fig. 4B). Importantly, GSEA also identified a significant dysfunction of hypoxia-inducible factor-1 (HIF-1) pathway after ALKBH1 depletion (Fig. 4C). The expression of critical genes such as HIF-1α, CA9, VEGFα, LDHA, PDK1, PDK4, and GLUT1 was significantly decreased, as shown in the heatmap (Fig. 4D). In addition, the downregulation of HIF-1 pathway was confirmed in Alkbh1-depleted 3T3-L1 cells by Western blot (Fig. 4E) and qPCR analysis (Fig. 4F). On the contrary, overexpression of ALKBH1 in 3T3-L1 cells significantly induced the protein level of HIF-1 axis (Fig. 4G) as well as the mRNA expression of Hif-1α, Ca9, Hk2, Vegfα, Glut1, and Ldha (Fig. 4H).
Figure 4.
ALKBH1 regulates HIF-1 pathway.A, Gene Ontology enrichment interprets the biological process in ALKBH1-knockdown hMSCs. Knockdown of ALKBH1 influenced the synthesis and transport of lipid. B, gene set enrichment analysis (GSEA) shows decreased expression of adipogenesis genes in ALKBH1-knockdown hMSCs. C, GSEA reveals that depletion of ALKBH1 in hMSCs impairs hypoxia pathway. D, heatmap of representative genes involved in HIF-1 signaling. E, Western blots of hypoxia-related proteins in Alkbh1-knocked 3T3-L1 cells in 20% oxygen conditions. F, qRT-PCR analysis of Hif-1α, Ca9, Hk2, Vegfα, Glut1, and Ldha in Alkbh1-deficient 3T3-L1 lines in 20% oxygen conditions (n = 3). G, Western blots and (H) qRT-PCR analysis confirm the activation of hypoxia axis in ALKBH1-overexpressed cells under normoxia (n = 3). I, qRT-PCR analysis of the mRNA levels of HIF-1 pathway. HIF-1 pathway activator ML228 upregulated related genes expression in control group but partially failed in Alkbh1-knockdown group (n = 3). The p values were calculated by two-tailed Student's t test (F and H) and one-way ANOVA with Tukey's post hoc test (I). Scatter plots show individual data points ± SD.
As HIF-1α is rapidly degraded by prolylhydroxylase (PHD) in normoxia (27), We cultured 3T3-L1 cells in 5% oxygen conditions. Knockdown of Alkbh1 led to a decreased expression of HIF-1α and its downstream targets both on protein level and mRNA level (Fig. S1, A and B), while overexpression of ALKBH1 activated the HIF-1 pathway (Fig. S1, C and D).
In addition, we introduced cobalt chloride (CoCl2), a canonical hypoxia mimic, to verify the results. We treated 3T3-L1 preadipocytes with 150 μM CoCl2 for 24 h to induce hypoxia, and the successful induction was confirmed by the elevation of Hif-1α (Fig. S2A). Consistent with our hypothesis, knockdown of Alkbh1 decreased the protein contents of HIF-1 targets (Fig. S2B) as well as gene expression of Ca9, Hk2, Vegfα, Glut1, and Ldha (Fig. S2C). Overexpression of ALKBH1 activated the HIF-1 pathway (Fig. S2, D and E).
In addition, we treated 3T3-L1 cells with the HIF-1 pathway agonist ML228, which successfully activated the HIF-1 pathway after 48 h of treatment (Fig. 4I). However, knockdown of Alkbh1 largely repressed this activation. A very mild increase of related genes was found in si-Alkbh1+ML228 group (Fig. 4I).
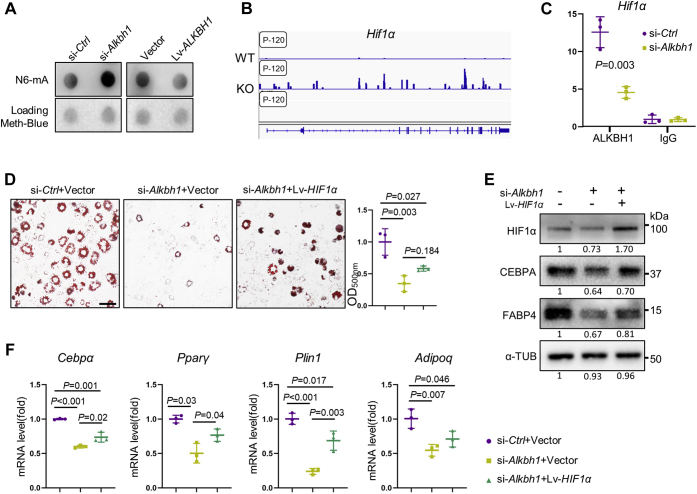
ALKBH1 demethylates DNA 6mA of HIF-1α
Next, we sought to assess if ALKBH1 functioned as a DNA 6mA demethylase by DNA dot blot assay. Depletion of Alkbh1 increased the 6mA level in whole genomic DNA, while overexpression of ALKBH1 reduced 6mA modifications in 3T3-L1 cells (Fig. 5A). By analyzing the published N6-mA-DNA-IP-seq dataset (GSE71866), we found that Hif-1α had a highly enriched and specific 6mA peak in Alkbh1-knockdown embryonic stem cells (ESCs) (Fig. 5B). Next, we confirmed that the binding of ALKBH1 at Hif-1α and its enrichment were reduced upon Alkbh1 depletion (Fig. 5C).
Figure 5.
ALKBH1 demethylates DNA 6mA of HIF-1α.A, levels of the DNA 6mA modification were assessed via DNA dot blot in 3T3-L1 cells using a 6mA-specific antibody. Methylene blue detected DNA loading. ALKBH1 functions as a DNA demethylase in 3T3-L1 cells. si-Ctrl: control-siRNA 3T3-L1 cells, si-Alkbh1: Alkbh1 deficient 3T3-L1 cells, Lv-ALKBH1: ALKBH1-overexpressed 3T3-L1 cells, Vector: Empty vector in 3T3-L1 cells. B, enrichment of N6-mA peak on Hif-1α gene locus in Alkbh1 knockout embryonic stem cells. Data are from the published datasets GSE71866. C, ChIP-PCR analysis of immunoprecipitations of ALKBH1 associated Hif-1α fragments. D, representative images of Oil red O staining in 3T3-L1 cells and quantitation analysis (n = 3). Scale bar: 25 μm. E, Western blot analysis of HIF-1α, CEBPA and FABP4. Overexpression of HIF-1α partially rescued the decrease of adipogenic markers. F, qRT-PCR shows the mRNA level of Cebpα, Pparγ, Plin1, and Adipoq in si-Ctrl+Vector, si-Alkbh1+Vector, and si-Alkbh1+Lv-HIF1α group at 5 days of differentiation (n = 3). The p values were calculated by two-tailed Student' t test (C) and one-way ANOVA with Tukey's post hoc test (D and F). Scatter plots show individual data points ± SD.
In addition, we infected 3T3-L1 cells with lentiviral particles overexpressing HIF-1α gene (Lv-HIF-1α), or empty vectors (Vector) and then knocked down Alkbh1 to perform the functional rescue experiments. Surprisingly, overexpression of HIF-1α only partially increased the maturity of lipid droplets compared with the knockdown group by oil red O staining (Fig. 5D). Besides, the protein contents of CEBPA and FABP4 received partial recovery (Fig. 5E), and mRNA levels of the key adipogenic genes Cebpα, Pparγ, Plin1, and Adipoq in the Lv-HIF1α+siAlkbh1 group were also partially restored after 5 days of induction (Fig. 5F), suggesting that ALKBH1 may regulate other genes.
Simultaneous overexpression of HIF-1α and GYS1 restores the adipogenic differentiation of Alkbh1-deficient cells
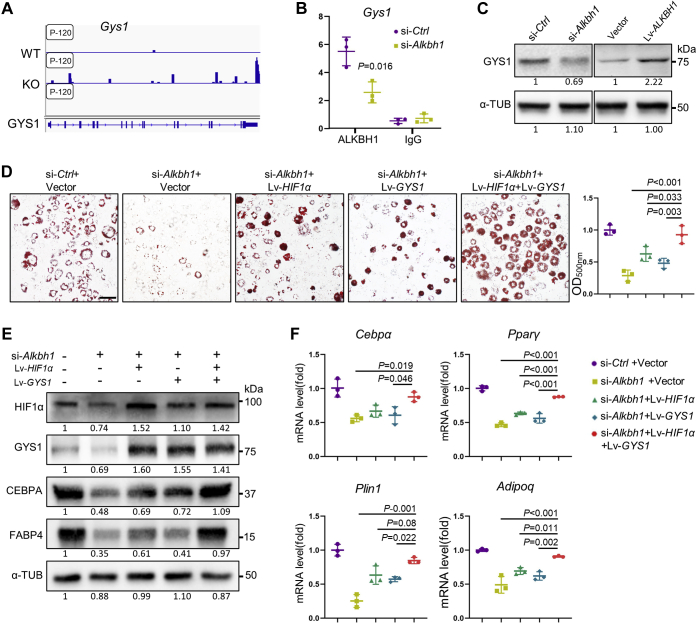
By overlapping our RNA-seq data with published N6-mA-DNA-IP-seq data (GSE71866), we also found that glycogen synthase 1 (Gys1), a downstream target of Hif-1α, exhibited a peak of DNA 6mA modification after the knockdown of Alkbh1 (Fig. 6A). The abundance of ALKBH1 at the promoter regions of Gys1 was reduced in Alkbh1-depleted 3T3-L1 cells (Fig. 6B). The protein contents of GYS1 were regulated by the level of ALKBH1 both in 20% oxygen conditions (Fig. 6C) and 5% oxygen (Fig. S3), suggesting that GYS1 may be another target of ALKBH1 in adipogenic differentiation.
Figure 6.
Simultaneous overexpression of HIF-1α and GYS1 restores the adipogenic differentiation of Alkbh1-deficient cells.A, DNA N6-mA peaks on Gys1 gene locus in Alkbh1 knockout embryonic stem cells. Data are from the published datasets GSE71866. B, ChIP-PCR analysis of immunoprecipitations of ALKBH1 associated Gys1 fragments (n = 3). C, Western blots of GYS1 in normoxia, indicating that ALKBH1 regulates the expression of GYS1. D, representative images and quantification of oil red O staining in si-Ctrl+Vector, si-Alkbh1+Vector, si-Alkbh1+Lv-HIF1α, si-Alkbh1+Lv-GYS1 group, and si-Alkbh1+Lv-HIF1α+GYS1 group (n = 3). Scale bar: 25 μm. E, Western blot detects the protein level of HIF-1α, GYS1 and adipogenic-related markers (n = 3). Simultaneous overexpression of HIF-1α and GYS1 restored adipogenic differentiation of Alkbh1-deficient cells. F, qRT-PCR analysis of the expression of Cebpα, Pparγ, Plin1, Adipoq after 5-day adipogenic differentiation (n = 3). The p values were calculated by two-tailed Student's t test (B) and one-way ANOVA with Tukey's post hoc test (D and F). Scatter plots show individual data points ± SD.
Overexpression of GYS1 alone failed to fully restore the decreased intensity of oil red O staining (Fig. S4A), protein contents of CEBPA and FABP4 (Fig. S4B), and mRNA level of adipogenic genes (Fig. S4C) in Alkbh1-depleted 3T3-L1 cells. Next, we simultaneously overexpressed HIF-1α and GYS1 and then knocked down Alkbh1 in those cells. Compared with cells that only overexpressed either of the two, amounts of lipid droplets in oil red O staining (Fig. 6D), as well as the adipogenic protein contents, were almost restored to their original level (Fig. 6E). Besides, the expression of adipogenesis-related genes Cebpα, Pparγ, Plin1, and Adipoq was significantly increased in cells concurrently overexpressing HIF-1α and GYS1 (si-Alkbh1+Lv-HIF1α+Lv-GYS1) after 5 days of adipogenic induction (Fig. 6F). In sum, ALKBH1 demethylated DNA 6mA of both Hif-1α and Gys1, which worked together to enhance adipogenic differentiation of 3T3-L1 cells.
Discussion
DNA modification, involving 5mC and 6mA in eukaryotes, is of paramount importance due to its extensive influence on embryonic development and metabolic homeostasis (28). Perturbation in 6mA leads to dysfunction and human diseases such as neurodegeneration (29), asymmetry (30), mitochondria disorder (31), and tumorigenesis (21, 32). In recent years, ALKBH1, which functions as a DNA 6mA demethylase, has played a versatile role in embryogenesis and differentiation. Loss of ALKBH1 results in defects in the nerves, skeleton, genitals, and cell pluripotency and differentiation (19, 25, 26). In the present study, we defined the relationship between ALKBH1 and cell adipogenic differentiation. Depletion of ALKBH1 in human MSCs and 3T3-L1 preadipocytes decreases adipogenesis. On the contrary, overexpression of ALKBH1 leads to lipid droplets accumulation in vitro and extensive adipose tissue in vivo. Combined with our previous data, ALKBH1 has a crucial role in cell fate determination (19).
We elucidated that ALKBH1 activates the HIF-1 signaling pathway to enhance adipogenesis in normoxia and hypoxia. HIF-1 consists of a functional α-subunit heterodimer and a stably expressed β-subunit in mammals (33). With the reduction of oxygen, HIF-1 is prevented from being hydroxylated by prolyl hydroxylase domain (PHD) and factor inhibiting HIF-1 (FIH1), thus promoting angiogenesis, epithelial-to-mesenchymal transition, and metastasis of tumors in eukaryotes (34, 35, 36). Aberrant level of HIF-1 leads to metabolic dysfunction such as mitochondrial diseases, β-cell disturbance, and glucose intolerance (37, 38). A growing number of studies suggest that hypoxia occurred during adipose accumulation in overweight individuals or obese mice due to increased oxygen consumption (39). Mice lacking HIF-1α present with a decrease of glycolytic enzymes (40), high insulin sensitivity, and less body weight after HFD diet (41, 42, 43, 44). While overexpression of HIF-1α in mice increases the obesity and insulin resistance interacting with inflammation and fibrosis of adipose tissue (45, 46, 47). The relationship between ALKBH1 and the HIF-1 pathway in tumorigenesis reported by Xiao et al. and Xie et al. (21, 32) also entrenched our hypothesis on adipogenesis. In recent years, studies paid more attention to the regulations of HIF-1α proteins due to their degradation under normoxia. The regulations of HIF-1α protein are well documented these years, whereas the transcription of DNA and regulation of mRNA is are little known. However, in many pathological conditions, such as in solid tumors and adipose tissue, HIF-1α is subjected to epigenetic regulations. The level of methylation on HIF-1α mediates the transcription and thus regulates the downstream targets. In this study, ALKBH1 knockdown reduced the global DNA methylation level of 6mA and attenuated the HIF-1 signaling pathway due to the hypermethylation of HIF-1α and GYS1. Hypermethylation of HIF-1α caused posttranscriptional gene silencing and consequently led to decreased adipogenic differentiation (44). Overexpression of HIF-1α could partially rescue the phenotypes. One explanation for this mechanism is that elevated expression of HIF-1α may activate insulin signaling and alter the level of adiponectin, a crucial gene that protects the body from metabolic diseases (48, 49). Controversial points existed due to the diversity of cell origins and imbalance of adipose inflammation (50, 51, 52). Besides, upstream genes of HIF-1α like ANT2 also contribute to obesity and inflammations (53, 54). However, whether ALKBH1 has direct effects on the degradation of HIF-1α needs further studies.
Glycogen synthase 1 (GYS1), downstream of HIF-1α, is the key enzyme functioning in the glycogen synthesis and significantly increases in the tumor microenvironment and hypoxic atmosphere (55, 56). Accumulation of GYS1 in several types of cancer suppresses canonical nuclear factor κB (NF-κB) signaling (57) and AMP-activated protein kinase (AMPK) signaling to promote migration and proliferation of tumor cells (58, 59, 60). Aberrant expression of GYS1 is also related to the obesity phenotype in diabetes, but the mechanism remains unknown. Here we found that GYS1 is also a target of ALKBH1. We speculated that GYS1 might further promote adipogenesis by altering gene expressions in NF-κB and AMPK signaling pathways.
ALKBH1 was discovered as a DNA 6mA demethylase to participate in epigenetic networks (20) and was shown to be a tRNA demethylase later on (23). He et al. reported that m1A58 tRNAiMet was the main substrate of ALKBH1 and can promote translational initiation. Recently, a number of studies uncovered the existence of 6mA in eukaryotes, indicating its function on cell differentiation (19), tumor progress (21, 32), and stress response (31). Of note, 6mA level in mtDNA arose under HIF1α activation. Accumulating evidence suggests that ALKBH1 can demethylase DNA 6mA, RNA m5C, m6A, and tRNA m1A because they share the locally unpairing feature (24).
HIF1α regulates physiological processes under both hypoxic and normoxic conditions (61, 62). Oxygen levels are highly heterogeneous among various tissues ranging from 1% to 5%, and controlled concentrations of oxygen help organs function normally (63). Hypoxia/oxygen-sensing signaling activates mainly regulated by HIFs and various enzymes. Moderate hypoxia mediated by HIFs regulates stem cell maintenance, differentiation, apoptosis, and mitochondrial functions at the cellular levels (64, 65, 66). But excessive hypoxia has side effects on metabolisms, angiogenesis, tissue fibrosis, inflammations, and tumor progression (67, 68). The discovery of novel mechanisms is critical for therapy. In summary, we uncover the crucial function of DNA 6mA modification on adipogenesis. The combination of two kinds of sequencings validates the relationship between ALKBH1 and hypoxia/HIF1α signaling for the first time and gives some new evidence on the regulations of adipogenesis.
Experimental procedures
Cell culture and adipogenic differentiation assays
Human bone-marrow-derived mesenchymal stem cells (hMSCs) and 3T3-L1 cell lines were obtained from ATCC. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-High Glucose (Hyclone laboratories, Logan, UT) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY), 100 μg/ml streptomycin (Gibco), and 100 units/ml penicillin (Gibco) at 37 °C in a humidified atmosphere of 5% CO2. To induce adipogenic differentiation, cells were seeded in 6-well plates and treated with adipogenic medium containing 0.5 μΜ 3-Isobutyl-1-methylxanthine (IBMX), 10 μg/ml insulin, 1 μM dexamethasone, and 72 μg/ml indomethacin (all from Sigma).
Gene knockdown and lentivirus-mediated gene overexpression
siRNAs for human ALKBH1 and control were purchased from Santa Cruz. Mouse siRNAs were synthesized by Sangon Biotech (Table S1). We diluted 1 μg siRNA duplex into 100 μl Opti-MEM I for solution A and diluted 5 μl Lipofectamine RNAiMAX (Invitrogen) into 100 μl Opti-MEM I for solution B. Then, we added A to B and incubated the mixture for 25 to 40 min at RT. When the cells reached 50 to 70% (about 1 × 105 cells) confluence, 200 μl mixture was added to 1 ml of normal growth medium without antibiotics in 6-well, incubating cells for 12 h at 37 °C. The knockdown efficiencies were examined by Western blot after 48 h of transfection.
For overexpression, the lentivirus-ALKBH1 vectors system was constructed, packaged, and purified by Genechem. Cells infected with an empty GV358 vectors (Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin) were used as controls. Cells were cultured in 6-well to 30% (about 5 × 104 cells) confluence and infected with lentivirus at a MOI = 20. The lentivirus solution along with 1 × Hitrans G Polybrene (Genechem) was added into 1 ml growth medium for 12 h at 37 °C. The green fluorescent could be observed after 72 h of infection. Then we used 2 μg/ml puromycin (Sigma) for 7 to 10 days to obtain stable cell clones. Cells overexpressing HIF-1α and GYS1 were also purchased from Genechem and infected in the same way. The efficiency of infection was confirmed by Western blots.
Cell viability
We used Cell Counting Kit-8 (CCK8, Beyotime) to check cell viability. Cells were treated with siRNA in 6-well and then digested after 24 h. Next, they were seeded and cultured at a density of 5 × 103/well into 96-well. After another 24, 48, and 72 h, 10 μl of CCK-8 reagent was added to each well and then cultured for 1 h. The absorbance was analyzed at 450 nm by a Spectrophotometer (Thermo Fisher Scientific).
qRT-PCR and RNA-Seq
We isolated total RNA by Trizol (Invitrogen) and reversed it to cDNA using PrimeScript RT reagent Kit and gDNA Eraser (TaKaRa Bio). Quantitative RT-PCR was performed by LightCycler 96 (Roche) with iTaq Universal SYBR Green Supermix (Bio-Rad laboratories). Genes of interest were calculated using a 2−ΔΔCt method by normalizing with RPLP0. All primer sequences are shown in Table S1.
For RNA-Seq, we prepared sequencing libraries using the Illumina TrueSeq stranded mRNA sample preparation kit (Illumina) according to the manufacturer's instruction and performed single-end sequences on an Illumina HiSeq 6000 machine. We used FastQC (v0.11.5) to control the quality of RNA-Seq and aligned the reads to hg19 genome using HISAT2 (v.0.0.5). Genes expression level was considered significantly changed if log2FC (Fold change) >0.5 or <−0.5, with p value <0.05 in our RNA-seq. GSEA for the related pathway was performed using GSEA software (http://www.broad.mit.edu/GSEA). Heat map in this study was drawn using pheatmap package in R 3.6.1 and normalized data by row (69).
Western blot
Cells were lysed by radioimmunoprecipitation assay (RIPA) buffer (Pierce) on ice to obtain total protein. Samples were centrifuged at 14,000g for 15 min at 4 °C and then heated at 95 °C for 5 min in 1× SDS loading buffer. We separated the protein by using 8% or 10% SDS–polyacrylamide gels and transferred it to PVDF membranes by a wet transfer apparatus (Bio-Rad). The membranes were blotted with 5% milk or bovine serum albumin (BSA) for 1 h and then incubated with primary antibodies at 4 °C overnight (70). The following antibodies were used: rabbit anti-ALKBH1 (1:2000, ab128895, Abcam), rabbit anti-HIF-1α (1:1000, ER1802-41, Huabio, Boston), rabbit anti-GYS1 (1:1000, ET1611-59, Huabio), rabbit anti-VEGF (1:2000, ET1604-28, Huabio), rabbit anti-GLUT1 (1:2000, ET1601-10, Huabio), rabbit anti-LDHA (1:2000, ET1608-57, Huabio), rabbit anti-CEBPA (1:2000, ET1612-46, Huabio), rabbit anti-FABP4 (1:2000, ET1703-98, Huabio), mouse anti-α-tubulin (1:2,000, sc-32293; Santa Cruz Biotechnology). The complexes were incubated in horseradish peroxidase conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Cell signaling Technology) and visualized with Immobilon reagents (Millipore).
Oil red O staining and Nile Red staining
Being inducted for 14 days, cells were fixed with 4% paraformaldehyde for 20 min, washed with PBS three times, and stained with Oil Red O (ORO) solution for 15 to 30 min (71). ORO stock solution was made with 0.5 g ORO (O8010, Solarbio) and 100 ml isopropyl alcohol. Six parts of saturated stock solution were dissolved in four parts of water to make the working fluid. Lipid droplets were visualized under the microscope (Olympus BX53).
Frozen tissue slices were managed following the tissue immunofluorescent staining procedure and then stained with Nile Red solution (N8440, Solarbio) for 10 min at 37 °C and DAPI before mounting. Images were captured by scanning confocal microscopy (FV3000, Olympus).
Ectopic fat formation
Nude mice were purchased from the Chengdu Dossy Experimental Animals CO. LTD. All animals were accommodated under 23 ± 2 °C and allowed for abundant food and water before injection. 2 × 105 3T3-L1 cells overexpressing ALKBH1 or empty vector mixed in 50% matrigel suspensions were respectively injected at different sides into the sternum of 4-week-old nude mice (1). Engrafted tissues were excised and histologically analyzed after 6 weeks of a high-fat diet. All animal studies performed were approved by the Subcommittee on Research and Animal Care (SRAC) of Sichuan University.
Histological evaluation
Fat tissues were harvested and fixed in the 4% paraformaldehyde (Biosharp) at 4 °C for 24 h and then dehydrated and embedded in paraffin (72). Samples were stained with hematoxylin and eosin (HE) following the manufacturer's instruction (Biosharp).
Tissue immunofluorescent staining
IWAT tissues were fixed in 4% paraformaldehyde (Biosharp) at 4 °C for 24 h, dehydrated in 30% sucrose solutions for 48 h, embedded with O.C.T compound (SAKURA) under −20 °C, and then sectioned at 5 μM (CM3050S; Leica). After three times of PBS and 1 h of blocking by 5% BSA, rabbit anti-GFP (ab290, Abcam) or anti-ALKBH1 primary antibody (1:2000, ab128895, Abcam) was applied at 4 °C overnight. Then secondary antibodies were applied at room temperature for 1.5 h by using anti-rabbit Alexa Fluor 488 (ab150077, Abcam). The nucleus was labeled by DAPI, and slides were captured by fluorescence microscope (Olympus BX53).
Dot-blot
We isolated DNA with a PureLink Genomic DNA kit (Invitrogen) and denatured it at 95 °C for 10 min in 0.4 M NaOH, 10 mM EDTA buffer. Samples were lightly spotted on the membrane (Zeta-Probe, Bio-Rad) using Dot-Blot microfiltration apparatus (Bio-Rad) and baked at 80 °C for 30 min. Membranes were blocked in 5% BSA for 1 h at room temperature, then incubated with N6-mA antibody (1:2000, 202–003, Synaptic Systems) overnight at 4 °C. The complexes were incubated with secondary anti-rabbit IgG (Jackson ImmunoResearch) for 1 h and visualized with Immobilon reagents (Millipore). To ensure an equal amount of DNA samples, the membrane was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) (19).
DNA-N6-mA-seq
The DNA-N6-mA-seq data used in this study were published in Wu et al. (20) with the accession number GSE71866. We loaded these data on Integrative Genomics Viewer (IGV), a high-performance desktop tool for interactive visual exploration of diverse genomic data, to compare the 6mA peak between the control and the knocked-Alkbh1 group (73).
Chromatin immunoprecipitation (ChIP)
The ChIP assay was conducted via an EZ-Zyme Chromatin Prep kit (Millipore) and Magna Chip HiSens (Millipore) (74). We use a rabbit IgG (Abcam) as control and a rabbit ALKBH1 (Abcam) for the experimental group. DNA–protein complexes were dissociated, and pulled-down DNA was tested by qRT-PCR analysis. The primers were designed to identify the target promoter regions. The ChIP assay results are demonstrated in relation to the input DNA.
Statistical analysis
Scatter plots show individual data points and standard deviation (SD). Statistically significant differences were performed by two-tailed Student's t test for comparison between two groups or by one-way analysis of variance (ANOVA) followed by the Tukey's post hoc test for multiple comparisons. p value <0.05 was considered to be statistically significant.
Data availability
The RNA-seq data have been uploaded and deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under GSE177059.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We appriciate Ms Xinyan Gan for assistance with language editing. This work was supported by grants from the Sichuan Science & Technology Program (2021YFH0015, 2020JDRC0023) and State Key Laboratory of Oral Diseases (SKLOD202111).
Author contributions
L. Y. and Y. Q. conceptualization; L. Y. and Lin Weimin formal analysis; Liu Weiqin and Y. Q. funding acquisition; L. Y., C. Y., J. S., and G. Y. investigation; L. Y., Wang Yuan, J. S., Wu Yunshu, and L. Q. methodology; Y. Q. project administration; L. Y., C. Y., and Y. Q. resources; L. Y. and Lin Weimin software; Y. Q. supervision; Y. Q. validation; L. Y. writing—original draft; L. Y., Wang Yuan, Wu Yunshu, L. Q., G. Y., and Liu Weiqin writing—review and editing.
Edited by Qi Qun Tang
Supporting information
References
- 1.Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schurmann A., Saraiva L.R., Schulz T.J. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20:771–784.e6. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin B., Yu F., Wang C., Li B., Liu M., Ye L. Epigenetic control of mesenchymal stem cell fate decision via histone methyltransferase Ash1l. Stem Cells. 2019;37:115–127. doi: 10.1002/stem.2918. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y., Li J., Sheng R., Deng P., Wang Y., Zheng R., Jiang Y., Ye L., Chen Q., Zhou X., et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai M., Devlin M.J., Rosen C.J. Fat targets for skeletal health. Nat. Rev. Rheumatol. 2009;5:365–372. doi: 10.1038/nrrheum.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cristancho A.G., Lazar M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Wang Y., Lin W., Sheng R., Wu Y., Xu R., Zhou C., Yuan Q. AFF1 inhibits adipogenic differentiation via targeting TGM2 transcription. Cell Prolif. 2020;53:e12831. doi: 10.1111/cpr.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Nie L., Zhao P., Zhou X., Ding Y., Chen Q., Wang Q. Diabetes fuels periodontal lesions via GLUT1-driven macrophage inflammaging. Int. J. Oral Sci. 2021;13:11. doi: 10.1038/s41368-021-00116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y., Zhang C., Lyu G., Huang X., Zhang X., Zhuang T., Jia L., Zhang L., Zhang C., Li C., Tao W. Senescence-activated enhancer landscape orchestrates the senescence-associated secretory phenotype in murine fibroblasts. Nucleic Acids Res. 2020;48:10909–10923. doi: 10.1093/nar/gkaa858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q., Wang M., Xue H., Liu W., Guo Y., Xu R., Shao B., Yuan Q. Ubiquitin-specific protease 34 inhibits osteoclast differentiation by regulating NF-kappaB signaling. J. Bone Miner. Res. 2020;35:1597–1608. doi: 10.1002/jbmr.4015. [DOI] [PubMed] [Google Scholar]
- 10.Skvortsova K., Iovino N., Bogdanovic O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018;19:774–790. doi: 10.1038/s41580-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Deng P., Liu Y., Wu Y., Chen Y., Guo Y., Zhang S., Zheng X., Zhou L., Liu W., Li Q., Lin W., Qi X., Ou G., Wang C., et al. Alpha-ketoglutarate ameliorates age-related osteoporosis via regulating histone methylations. Nat. Commun. 2020;11:5596. doi: 10.1038/s41467-020-19360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez-Errico D., Vento-Tormo R., Sieweke M., Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 2015;15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 13.Daniunaite K., et al. Epigenetic regulation of human adipose-derived stem cells differentiation. Mol. Cell. Biochem. 2015;410:111–120. doi: 10.1007/s11010-015-2543-7. [DOI] [PubMed] [Google Scholar]
- 14.Xu Q., Xie W. Epigenome in early mammalian development: Inheritance, reprogramming and establishment. Trends Cell Biol. 2018;28:237–253. doi: 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich M. DNA hypermethylation in disease: Mechanisms and clinical relevance. Epigenetics. 2019;14:1141–1163. doi: 10.1080/15592294.2019.1638701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Silva T.C., Young J.I., Gomez L., Schmidt M.A., Hamilton-Nelson K.L., Kunkle B.W., Chen X., Martin E.R., Wang L. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer's disease. Nat. Commun. 2020;11:6114. doi: 10.1038/s41467-020-19791-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Q., Wang C., Kuang X., Feng X., Sartorelli V., Ying H., Ge K., Dent S.Y. Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis. Mol. Cell. Biol. 2014;34:3746–3753. doi: 10.1128/MCB.00622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C., Liu Y., Li X., Zou J., Zou S. DNA N6-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016;4:16033. doi: 10.1038/boneres.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M., Tackett A., Wang G., Hon L.S., Fang G., Swenberg J.A., et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao C.L., Zhu S., He M., Chen, Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F., Liang Z., Wang D.P., Bo X.C., Gu X.F., Wang K., et al. N(6)-Methyladenine DNA modification in the human genome. Mol. Cell. 2018;71:306–318.e7. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Ougland R., Lando D., Jonson I., Dahl J.A., Moen M.N., Nordstrand L.M., Rognes T., Lee J.T., Klungland A., Kouzarides T., Larsen E. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012;30:2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G., Ma H., Han D., Evans M., Klungland A., Pan T., et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M., Yang S., Nelakanti R., Zhao W., Liu G., Li Z., Liu X., Wu T., Xiao A., Li H. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020;30:197–210. doi: 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Z., Sikandar S., Witherspoon M., Dizon D., Nguyen T., Benirschke K., Wiley C., Vrana P., Lipkin S.M. Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice. Dev. Dyn. 2008;237:316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- 26.Nordstrand L.M., Svard J., Larsen E., Nilsen A., Ougland R., Furu K., Lien G.F., Rognes T., Namekawa S.H., Lee J.T., Klungland A. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi Y., Yokota A., Harada H., Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1alpha in cancer. Cancer Sci. 2019;110:1510–1517. doi: 10.1111/cas.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 29.Yao B., Cheng Y., Wang Z., Li Y., Chen L., Huang L., Zhang W., Chen D., Wu H., Tang B., Jin P. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017;8:1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Liu Z., Lin H., Ma D., Tao Q., Liu F. Epigenetic regulation of left-right asymmetry by DNA methylation. EMBO J. 2017;36:2987–2997. doi: 10.15252/embj.201796580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma C., Niu R., Huang T., Shao L.W., Peng Y., Ding W., Wang Y., Jia G., He C., Li C.Y., He A., Liu Y. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019;21:319–327. doi: 10.1038/s41556-018-0238-5. [DOI] [PubMed] [Google Scholar]
- 32.Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U., Zhang C., Dowiak A.V., Lin K., Zeng C., Sui Y., et al. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175:1228–1243.e20. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakker W.J., Harris I.S., Mak T.W. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol. Cell. 2007;28:941–953. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari A., Tashiro K., Dixit A., Soni A., Vogel K., Hall B., Shafqat I., Slaughter J., Param N., Le A., Saunders E., Paithane U., Garcia G., Campos A.R., Zettervall J., et al. Loss of HIF1A from pancreatic cancer cells increases expression of PPP1R1B and degradation of p53 to promote invasion and metastasis. Gastroenterology. 2020;159:1882–1897.e5. doi: 10.1053/j.gastro.2020.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 36.Nordgren I.K., Tavassoli A. Targeting tumour angiogenesis with small molecule inhibitors of hypoxia inducible factor. Chem. Soc. Rev. 2011;40:4307–4317. doi: 10.1039/c1cs15032d. [DOI] [PubMed] [Google Scholar]
- 37.Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., Hsu K.W., Lin Y.T., Peng P.H., Zhang L.S., Gao Y., Hu L., Sun H.L., Zhu A., Liu J., et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell. 2020;78:382–395.e8. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes R.A., Cheng K., Deters N., Lau S.M., Hawthorne W.J., O'Connell P.J., Stolp J., Grey S., Loudovaris T., Kay T.W., Thomas H.E., Gonzalez F.J., Gunton J.E. Hypoxia-inducible factor-1alpha (HIF-1alpha) potentiates beta-cell survival after islet transplantation of human and mouse islets. Cell Transpl. 2013;22:253–266. doi: 10.3727/096368912X647180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.S., Kim J.W., Osborne O., Oh D.Y., Sasik R., Schenk S., Chen A., Chung H., Murphy A., Watkins S.M., Quehenberger O., Johnson R.S., Olefsky J.M. Increased adipocyte O2 consumption triggers HIF-1alpha, causing inflammation and insulin resistance in obesity. Cell. 2014;157:1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basse A.L., Isidor M.S., Winther S., Skjoldborg N.B., Murholm M., Andersen E.S., Pedersen S.B., Wolfrum C., Quistorff B., Hansen J.B. Regulation of glycolysis in brown adipocytes by HIF-1alpha. Sci. Rep. 2017;7:4052. doi: 10.1038/s41598-017-04246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jun J.C., Devera R., Unnikrishnan D., Shin M.K., Bevans-Fonti S., Yao Q., Rathore A., Younas H., Halberg N., Scherer P.E., Polotsky V.Y. Adipose HIF-1alpha causes obesity by suppressing brown adipose tissue thermogenesis. J. Mol. Med. (Berl) 2017;95:287–297. doi: 10.1007/s00109-016-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trayhurn P., Wang B., Wood I.S. Hypoxia in adipose tissue: A basis for the dysregulation of tissue function in obesity? Br. J. Nutr. 2008;100:227–235. doi: 10.1017/S0007114508971282. [DOI] [PubMed] [Google Scholar]
- 43.Matsuura H., Ichiki T., Inoue E., Nomura M., Miyazaki R., Hashimoto T., Ikeda J., Takayanagi R., Fong G.H., Sunagawa K. Prolyl hydroxylase domain protein 2 plays a critical role in diet-induced obesity and glucose intolerance. Circulation. 2013;127:2078–2087. doi: 10.1161/CIRCULATIONAHA.113.001742. [DOI] [PubMed] [Google Scholar]
- 44.Jiang C., Qu A., Matsubara T., Chanturiya T., Jou W., Gavrilova O., Shah Y.M., Gonzalez F.J. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes. 2011;60:2484–2495. doi: 10.2337/db11-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regazzetti C., Peraldi P., Gremeaux T., Najem-Lendom R., Ben-Sahra I., Cormont M., Bost F., Le Marchand-Brustel Y., Tanti J.F., Giorgetti-Peraldi S. Hypoxia decreases insulin signaling pathways in adipocytes. Diabetes. 2009;58:95–103. doi: 10.2337/db08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halberg N., Khan T., Trujillo M.E., Wernstedt-Asterholm I., Attie A.D., Sherwani S., Wang Z.V., Landskroner-Eiger S., Dineen S., Magalang U.J., Brekken R.A., Scherer P.E. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol. Cell. Biol. 2009;29:4467–4483. doi: 10.1128/MCB.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takikawa A., Mahmood A., Nawaz A., Kado T., Okabe K., Yamamoto S., Aminuddin A., Senda S., Tsuneyama K., Ikutani M., Watanabe Y., Igarashi Y., Nagai Y., Takatsu K., Koizumi K., et al. HIF-1alpha in myeloid cells promotes adipose tissue remodeling toward insulin resistance. Diabetes. 2016;65:3649–3659. doi: 10.2337/db16-0012. [DOI] [PubMed] [Google Scholar]
- 48.Jiang C., Kim J.H., Li F., Qu A., Gavrilova O., Shah Y.M., Gonzalez F.J. Hypoxia-inducible factor 1alpha regulates a SOCS3-STAT3-adiponectin signal transduction pathway in adipocytes. J. Biol. Chem. 2013;288:3844–3857. doi: 10.1074/jbc.M112.426338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanatani Y., Usui I., Ishizuka K., Bukhari A., Fujisaka S., Urakaze M., Haruta T., Kishimoto T., Naka T., Kobayashi M. Effects of pioglitazone on suppressor of cytokine signaling 3 expression: Potential mechanisms for its effects on insulin sensitivity and adiponectin expression. Diabetes. 2007;56:795–803. doi: 10.2337/db06-1039. [DOI] [PubMed] [Google Scholar]
- 50.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 51.Saltiel A.R., Olefsky J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 2017;127:1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao M., Hepler C., Zhang Q., Shan B., Vishvanath L., Henry G.H., Zhao S., An Y.A., Wu Y., Strand D.W., Gupta R.K. Pathologic HIF1alpha signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell. 2021;28:685–701.e7. doi: 10.1016/j.stem.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leake I. ANT2 mediates hypoxia and inflammation in obesity. Nat. Rev. Endocrinol. 2019;15:64. doi: 10.1038/s41574-018-0140-z. [DOI] [PubMed] [Google Scholar]
- 54.Seo J.B., Riopel M., Cabrales P., Huh J.Y., Bandyopadhyay G.K., Andreyev A.Y., Murphy A.N., Beeman S.C., Smith G.I., Klein S., Lee Y.S., Olefsky J.M. Knockdown of Ant2 reduces adipocyte hypoxia and improves insulin resistance in obesity. Nat. Metab. 2019;1:86–97. doi: 10.1038/s42255-018-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Favaro E., Bensaad K., Chong M.G., Tennant D.A., Ferguson D.J., Snell C., Steers G., Turley H., Li J.L., Gunther U.L., Buffa F.M., McIntyre A., Harris A.L. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 2012;16:751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Pelletier J., Bellot G., Gounon P., Lacas-Gervais S., Pouyssegur J., Mazure N.M. Glycogen synthesis is induced in hypoxia by the hypoxia-inducible factor and promotes cancer cell survival. Front Oncol. 2012;2:18. doi: 10.3389/fonc.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S.L., Huang Q.S., Huang Y.H., Yang X., Yang M.M., He Y.F., Cao Y., Guan X.Y., Yun J.P. GYS1 induces glycogen accumulation and promotes tumor progression via the NF-kappaB pathway in clear cell renal carcinoma. Theranostics. 2020;10:9186–9199. doi: 10.7150/thno.46825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimomura H., Sanke T., Ueda K., Hanabusa T., Sakagashira S., Nanjo K. A missense mutation of the muscle glycogen synthase gene (M416V) is associated with insulin resistance in the Japanese population. Diabetologia. 1997;40:947–952. doi: 10.1007/s001250050772. [DOI] [PubMed] [Google Scholar]
- 59.Xirouchaki C.E., Mangiafico S.P., Bate K., Ruan Z., Huang A.M., Tedjosiswoyo B.W., Lamont B., Pong W., Favaloro J., Blair A.R., Zajac J.D., Proietto J., Andrikopoulos S. Impaired glucose metabolism and exercise capacity with muscle-specific glycogen synthase 1 (gys1) deletion in adult mice. Mol. Metab. 2016;5:221–232. doi: 10.1016/j.molmet.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pederson B.A., Schroeder J.M., Parker G.E., Smith M.W., DePaoli-Roach A.A., Roach P.J. Glucose metabolism in mice lacking muscle glycogen synthase. Diabetes. 2005;54:3466–3473. doi: 10.2337/diabetes.54.12.3466. [DOI] [PubMed] [Google Scholar]
- 61.Wielockx B., Grinenko T., Mirtschink P., Chavakis T. Hypoxia pathway proteins in normal and malignant hematopoiesis. Cells. 2019;8:155. doi: 10.3390/cells8020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaelin W.G., Jr., Ratcliffe P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 63.Schodel J., Ratcliffe P.J. Mechanisms of hypoxia signalling: New implications for nephrology. Nat. Rev. Nephrol. 2019;15:641–659. doi: 10.1038/s41581-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 64.Trayhurn P. Hypoxia and adipocyte physiology: Implications for adipose tissue dysfunction in obesity. Annu. Rev. Nutr. 2014;34:207–236. doi: 10.1146/annurev-nutr-071812-161156. [DOI] [PubMed] [Google Scholar]
- 65.Semenza G.L. Hypoxia-inducible factor 1: Regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta. 2011;1813:1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee K.E., Simon M.C. From stem cells to cancer stem cells: HIF takes the stage. Curr. Opin. Cell Biol. 2012;24:232–235. doi: 10.1016/j.ceb.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Majmundar A.J., Wong W.J., Simon M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu K. Become competent in generating RNA-seq Heat maps in one day for novices without prior R experience. Methods Mol. Biol. 2021;2239:269–303. doi: 10.1007/978-1-0716-1084-8_17. [DOI] [PubMed] [Google Scholar]
- 70.Jiang S., Sheng R., Qi X., Wang J., Guo Y., Yuan Q. USP34 regulates tooth root morphogenesis by stabilizing NFIC. Int. J. Oral Sci. 2021;13:7. doi: 10.1038/s41368-021-00114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y., Yuan Q., Vergnes L., Rong X., Youn J.Y., Li J., Yu Y., Liu W., Cai H., Lin J.D., Tontonoz P., Hong C., Reue K., Wang C.Y. KDM4B protects against obesity and metabolic dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5566–E5575. doi: 10.1073/pnas.1721814115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang D., Zhang S., Wang J., Li Q., Xue H., Sheng R., Xiong Q., Qi X., Wen J., Fan Y., Zhou B.O., Yuan Q. LepR-expressing stem cells are essential for alveolar bone regeneration. J. Dent. Res. 2020;99:1279–1286. doi: 10.1177/0022034520932834. [DOI] [PubMed] [Google Scholar]
- 73.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Xiao Q., Wu Z., Xu R., Zou S., Zhou C. AFF4 enhances odontogenic differentiation of human dental pulp cells. Biochem. Biophys. Res. Commun. 2020;525:687–692. doi: 10.1016/j.bbrc.2020.02.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been uploaded and deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under GSE177059.