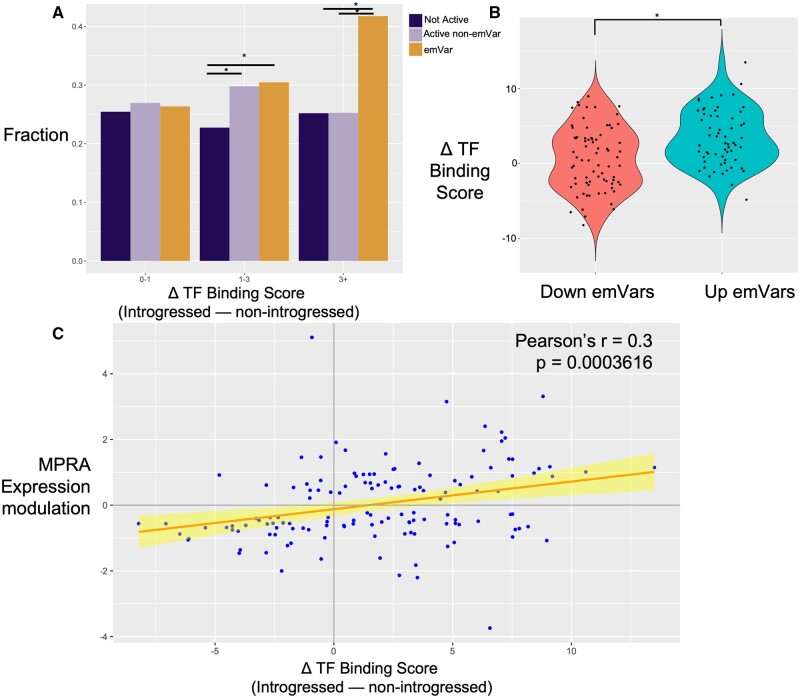
Fig. 3.
Transcription factor-binding site analysis of emVars. (A) After curating a set of predicted TF-binding motifs in K562 cells (see Materials and Methods) and examining at each introgressed and nonintrogressed sequence how each variant alters binding strength (deltaTF Binding score), we compared different classes of variants (not-active variants, active non-emVars, and emVars) for their overall impacts on differential binding. In general, emVars are more likely to lead to both large changes (3+) in predicted binding strength than either nonactive sequences (OR=1.91, P = 2.1×10−5) or active sequences with no emVar (OR=1.93, P = 2.9×10−5). emVars and active sequences with no emVar are both more likely than nonactive sequences to have moderate changes (1–3) in predicted binding strength (OR emVar vs. nonactive=1.42, P = 2.4×10−5, OR active vs. nonactive=1.36, P = 0.002). (B) We then compared binding affinities at each variant in each emVar to their direction of effect on expression in MPRA. In general, emVars that upregulated expression in the MPRA were predicted to have stronger binding to TFs than those that downregulated in the MRPA (t-test P = 6.9×10−5). (C) Correlation analysis between emVar MPRA expression modulating effect (LFC cDNA:pDNA) and the predicted change in TF-binding score, noting a significant moderate correlation between (r = 0.3, P = 0.0004). In all three panels, delta (Δ) TF-binding score is calculated as the introgressed TF-binding score minus the nonintrogressed binding score.