Figure 2: Image registration using background signal is similar to that obtained using DAPI.
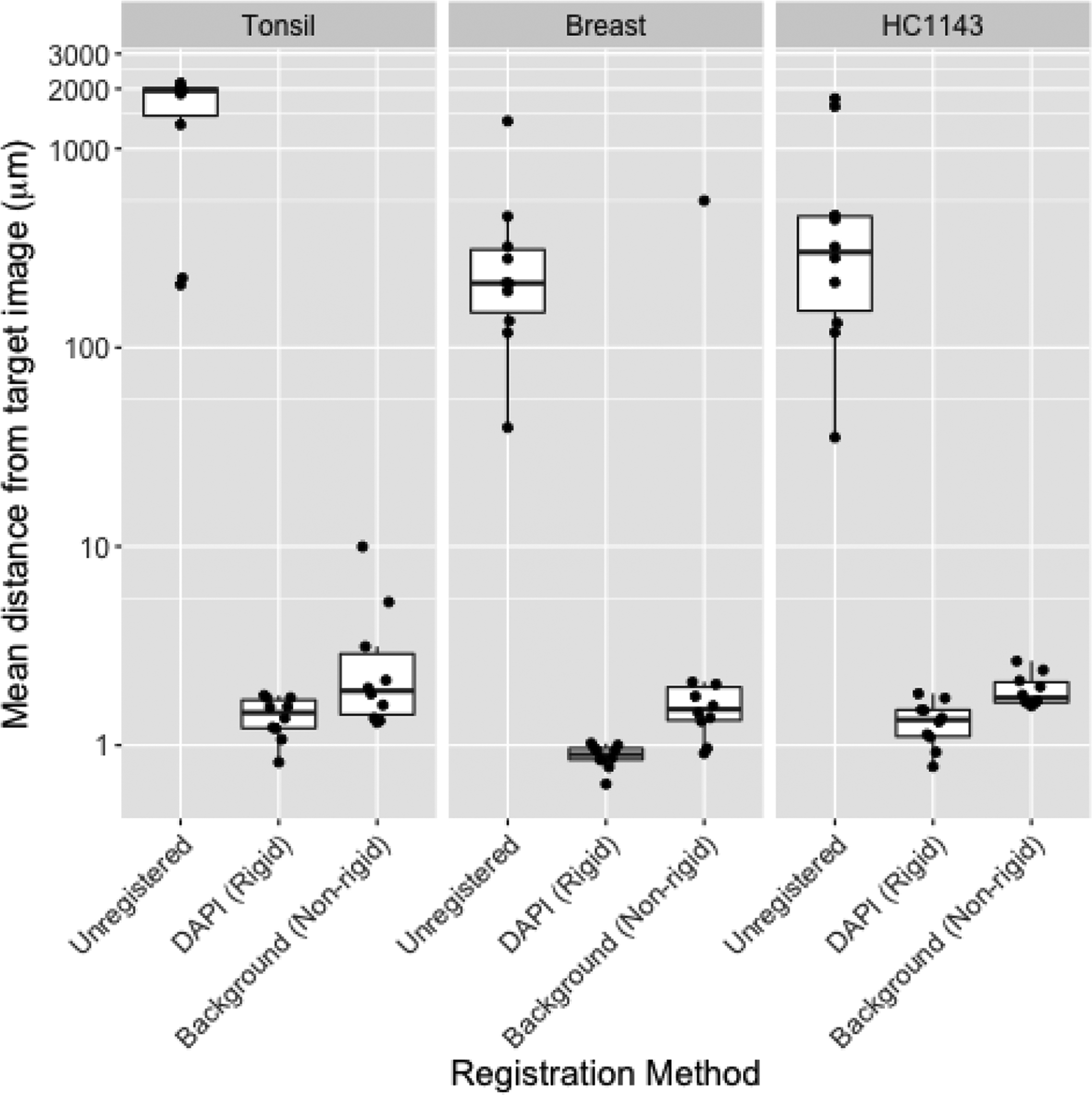
Mean distance from round 1 image (R1; y axis) to images in all other rounds (n=10) before registration (“Unregistered”), following rigid registration using the DAPI channel [“DAPI (Rigid)”], or following a non-rigid registration using the background channel [“Background (Non-rigid)”] in tissue microarray images from healthy tonsil tissue (left), healthy breast tissue (center), or HC1143 breast cancer cell line (right). Distance is calculated by defining AKAZE (Alcantarilla et al., 2013) keypoints in both images, matching the keypoints across images, summarizing the distance between matched keypoints using target registration error (TRE) (Maurer et al., 1997), and scaling TRE (whose denominator is the diagonal of the image in pixels) by the length of the image diagonal in microns. Non-rigid registration was performed using SimpleElastix (Marstal et al., 2016); rigid registration was performed using AKAZE-based keypoint matching in OpenCV (Bradski, 2000).
