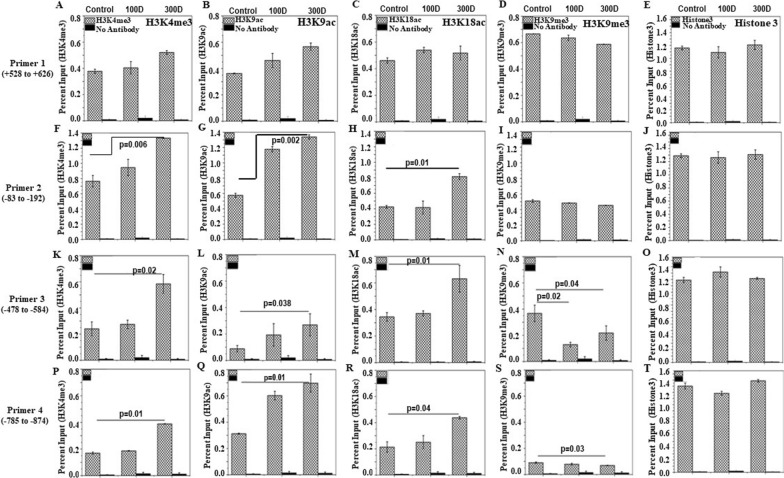
Fig. 4.
ChIP analysis of histone H3 methylation and acetylation at the region covering upstream to downstream of the promoter of the securin gene. ChIP-qRT-PCR assays for H3K4me3, H3K9ac, H3K18ac and H3K9me3 recruitment to the four regions of the securin gene + 528 to + 626 (upper panel A–E), − 83 to − 192 (F–J), − 478 to − 584 (K–O) and -785 to -874 (lower panel P–T) were analysed by 4 sets of primers in mouse stomach cells treated with RAN + lime for 0, 100 and 300 days. Chromatin was cross-linked, fragmented and immunoprecipitated with no antibody (as a negative control), with Histone 3 antibody (as a positive control) or anti-H3K4me3, H3K9ac, H318ac and H3K9me3 ChIP-grade antibodies. The purified DNA was used to amplify with four sets of primer pairs covering four regions (+ 528 to + 626 with Primer 1; − 83 to − 192 with Primer 2; − 478 to − 584 with Primer 3 and − 785 to − 874 with Primer 4) of the securin promoter by qPCR. As input, 10% diluted chromatin fragments were retained and used in qPCR for the enrichment analysis. The percentage of input values represents the mean of four different animals ± SEM. Data were analysed using one-way ANOVA with Tukey’s multiple comparison post-tests. P values less than 0.05 are considered significant