Abstract
Designer nanomaterials capable of delivering immunomodulators to specific immune cells have been extensively studied. However, emerging evidence suggests that several of these nanomaterials can non-specifically activate NLRP3 inflammasomes, an intracellular multiprotein complex controlling various immune cell functions, leading to undesirable effects. To understand what nanoparticle-attributes activate inflammasomes, we designed a multiparametric polymer supramolecular nanoparticle system to modulate various surface and core Nanoparticle-Associated Molecular Patterns (NAMPs), one at a time. We also investigated several underlying signaling pathways, including lysosomal rupture-cathepsin B maturation and calcium flux-mitochondrial ROS production, to gain mechanistic insights into NAMPs mediated inflammasome activation. Here we report that out of the four NAMPs tested, core hydrophobicity strongly activates and positively correlates with the NLRP3 assembly compared to surface charge, core rigidity and surface hydrophobicity. Moreover, we demonstrate different signaling inclinations and kinetics followed by differential core hydrophobicity patterns with the most hydrophobic ones exhibiting both lysosomal rupture and calcium influx early on. Altogether, this study will help design the next generation of polymeric nanomaterials for specific regulation of inflammasome activation, aiding efficient immunotherapy and vaccine delivery.
Graphical Abstract
INTRODUCTION
The inflammasome is a hetero-multimeric protein complex known for activating inflammatory caspases followed by subsequent processing of prointerleukin-1β which makes it one of the key players during inflammation. (1) This high molecular weight protein complex gets assembled and activated in response to microbial invasion or cellular damage detected mainly by phagocytic innate immune cells like macrophages and dendritic cells. These pathogenic interventions or tissue injury generate pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), respectively, that can be sensed by a myriad of threat sensors, classified into surface TLRs (Toll-like receptors) and cytosolic NLRs (NOD-like receptors). (2) Based on the type of stimuli and the responding NLRs, the inflammasome class is decided. NLRP3 inflammasome has been widely studied due to its broad stimuli range resulting in its involvement in various inflammatory diseases. (3–9) NLRP3 complex assembly and activation require two signals: priming, initiated by TLR4 agonists or inflammatory cytokines leading to activation of NFκB transcription factor; and activation, triggered by DAMPs causing cellular damage activity like lysosomal disruption, calcium influx, and mitochondrial ROS production. The inflammasome complex components (NLRP3 threat sensor, ASC adaptor and pro-caspase-1 effector enzyme) generated during priming get assembled after signal two, leading to maturation of the pro-caspase-1. The mature caspase-1 then acts upon the downstream inactive proteins (inactive gasdermin and cytokines) and cleaves at specific substrates to liberate their active forms. Active Gasdermin D further interacts with the cell plasma membrane to induce pore formation, ultimately causing cell lysis, popularly known as pyroptosis. It is associated with the release of active inflammatory cytokines IL-1β and IL-18, leading to enhanced immune response.
Several studies have investigated nanoparticle interactions with immune cells to understand their role in various biological applications and tailor them to different needs. (10–15) However, only a handful of studies have investigated this in the context of inflammasome activation, leaving unexplored prospects. Most of these studies have focused on identifying and reporting different inflammasome-activating nanoparticles like SiO2, TiO2, mesoporous silica, iron oxide, silver, carbon black and DOTAP particles (16–35). In contrast, not enough attention has been paid to a comprehensive characterization of Nanoparticle-Associated-Molecular-Patterns (NAMPs) responsible for it. Studies to date have reported only a few nanoparticle-attributes responsible for mediating inflammasome activation in the above-listed nano-platforms (31–33, 35–43) with very little focus on polymeric nanoparticles (23, 36–38) due to a lack of appropriate investigative tools. This renders an incomplete picture of different NAMPs stimulating NLRP3 assembly and their interplay in a single polymeric nanoplatform, leaving the prospect for identifying novel NAMPs and associated signaling cascades. Moreover, core nanoparticle attributes have been largely unexplored in the previous studies; despite having an important role in other immune interactions, eventually stimulating cytokines like TNFα and IFNγ. (38, 44–47) Nanoparticle surface and core hydrophobicity have also been shown to influence membrane permeability and endo-lysosomal escape or rupture (NLRP3 signal 2),(38, 44, 48, 49), making them potential candidates for investigation.
Here, we proposed a comprehensive study covering multiple surface and core NAMPs in a single polymeric supramolecular nanoparticle system, tweaking one nanoparticle property at a time to allow us to test numerous NAMPs regulating inflammasome activation using one platform. Simultaneously, we examined multiple associated cellular damage pathways to gain complete insights into the mechanisms underlying these activation events. In total, we tested four series of nanoparticles representing different NAMPs, including surface charge, surface hydrophobicity, core rigidity and core hydrophobicity. We first started with examining the effects of varying surface charge on NLRP3 assembly and activation as it is the first component interacting with the cell surface, we next examined surface hydrophobicity as it is well known to dictate immune responses but poorly studied in inflammasome context. We further tested core rigidity as it has been shown to influence the cell surface permeability important for changing flux responses, one of the plausible reasons for inflammasome activation.(50) Using this supramolecular nanoparticle system, we observed that surface properties, including charge and hydrophobicity, are insufficient to activate the NLRP3 complex even with a rigid core structure.
Interestingly, our findings also reveal core hydrophobicity as a novel NAMP regulating NLRP3 activation via more than one signaling cascade, including lysosomal rupture-cathepsin B and calcium flux-mitochondrial ROS production arrays (Figure 1). To further characterize this novel NAMP, we engineered the polymeric supramolecular nanoparticle system to obtain varying degrees of core hydrophobicity, which enabled us to identify its direct correlation with NLRP3 assembly and peek into their respective signaling trends as well as kinetics. Overall, this is the first study utilizing a designer polymeric supramolecular nanoparticle system that offers precise control over its several surface and core NAMPs to determine their impact on NLRP3 assembly and activation. A comprehensive analysis of different NAMPs regulating inflammasome activation in a polymer system will help select a safer biomaterial for delivery and assist in modulating nanosystems to achieve controlled inflammasome activation. We anticipate that this study will guide in establishing the groundwork for identifying therapeutically relevant nano-systems that can aid in immunotherapy and vaccine delivery (51–53).
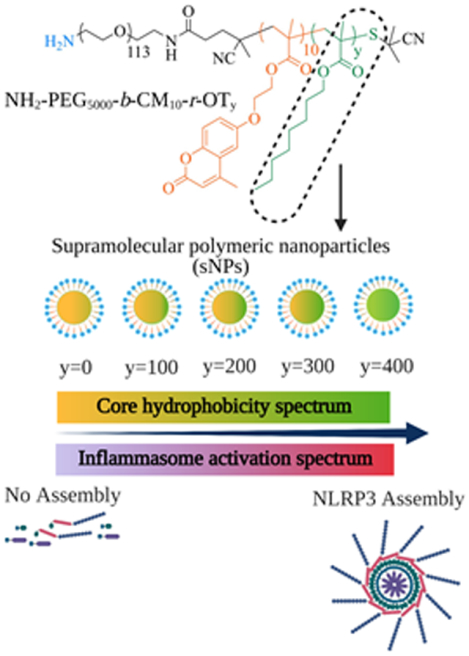
Figure 1. Schematic illustration of core hydrophobicity as a potential Nanoparticle-Associated-Molecular Pattern (NAMP) responsible for activating NLRP3 inflammasome and its proposed mode of action.
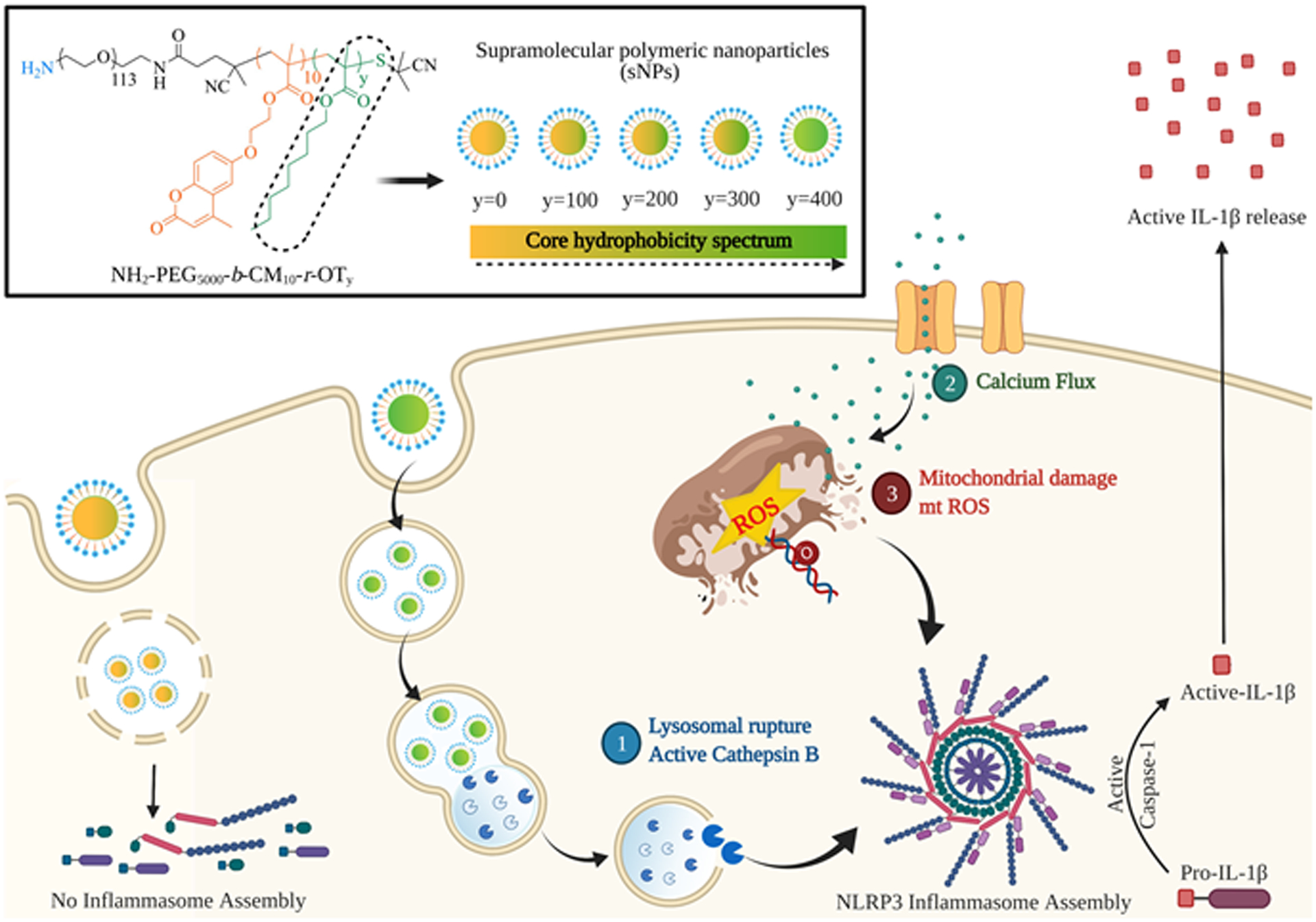
Top part shows the structures of the polymer construct used for the synthesis of supramolecular nanoparticles (sNPs) with sequential core hydrophobicity contributed by octyl hydrophobic moiety (y). The bottom part displays their detailed signaling pathway for NLRP3 inflammasome activation.
RESULTS
Synthesis and characterization of supramolecular nanoparticles (sNPs) with varying NAMPs.
To synthesize different series of sNPs, distinct variations are introduced into a common parent polymeric backbone, “R-PEG5000-b-CM10” (Figure 2A). The first series (series 1) of sNPs consists of varying surface charges, as a result of a distinct “R” group in the R-PEG5000-b-CM10 polymer, represented as R1, R2 and R3 (Figure 2A). R(1)-PEG5000-b-CM10, R(2)-PEG5000-b-CM10 and R(3)-PEG5000-b-CM10 are utilized to formulate positively, negatively, and neutral surface charged nanoparticles. Positively charged R(1)-PEG5000-b-CM10 polymer was synthesized as per the previous procedure. (54) Neutral charged R(3)-PEG5000-b-CM10 and negatively charged R(2)-PEG5000-b-CM10 polymers were synthesized according to scheme S1 and S2, respectively. Nitrophenylcarbamate group was used as the neutral moiety since, in our previous studies, it was demonstrated that it remains in the neutral condition after nanoparticle synthesis and gets converted to charged moieties upon UV irradiation.(55) However, in this study, since we didn’t use UV irradiation to active the functional group, the nitrophenylcarbamate group was used as a neutral functional group that was insufficient to induce inflammasome activation. The second series (series 2) of sNPs involved all three differently charged particles with a rigid core. This rigidity was introduced by crosslinking the coumarin (Cum, CM) moiety of the respective polymers. To attain these, sNPs from series 1 were irradiated with 365 nm light for 3 minutes in a UV incubator. The crosslinked nanoparticles with higher core rigidity were termed as R(1)-PEG5000-b-CM10XL R(2)-PEG5000-b-CM10XL and R(3)-PEG5000-b-CM10XL, with XL denoting crosslinked (Figure 2A). Third series (series 3) of sNPs included particles with the surface hydrophobic patch, obtained from R(4)-PEG5000-b-CM10 polymeric variant (Figure 2A). Fourth series (series 4) sNPs consisted of varying core hydrophobicity, contributed by different units (y) of the octyl hydrophobic moiety (Oct). We started with the highest hydrophobic particles comprising of 400 Oct units and then extended the series to acquire different y values (0 to 300) representing differential core hydrophobicity. In total, this series of nanoparticles were synthesized from NH2-PEG5000-b-CM10-r-OTy polymer, where “y” varied from 0 to 400, with “0” denoting least and “400” indicating most hydrophobic core. The range of 0 to 400 was selected as they efficiently formed monodispersed sNPs required for later steps. Further, these series of polymers were synthesized via multistep reactions as outlined in scheme S3. All polymers were characterized by 1H and 13C NMR (Supplementary Materials, section 2).
Figure 2. Screening and identification of NAMPs responsible for inflammasome activation by measuring IL-1β release.
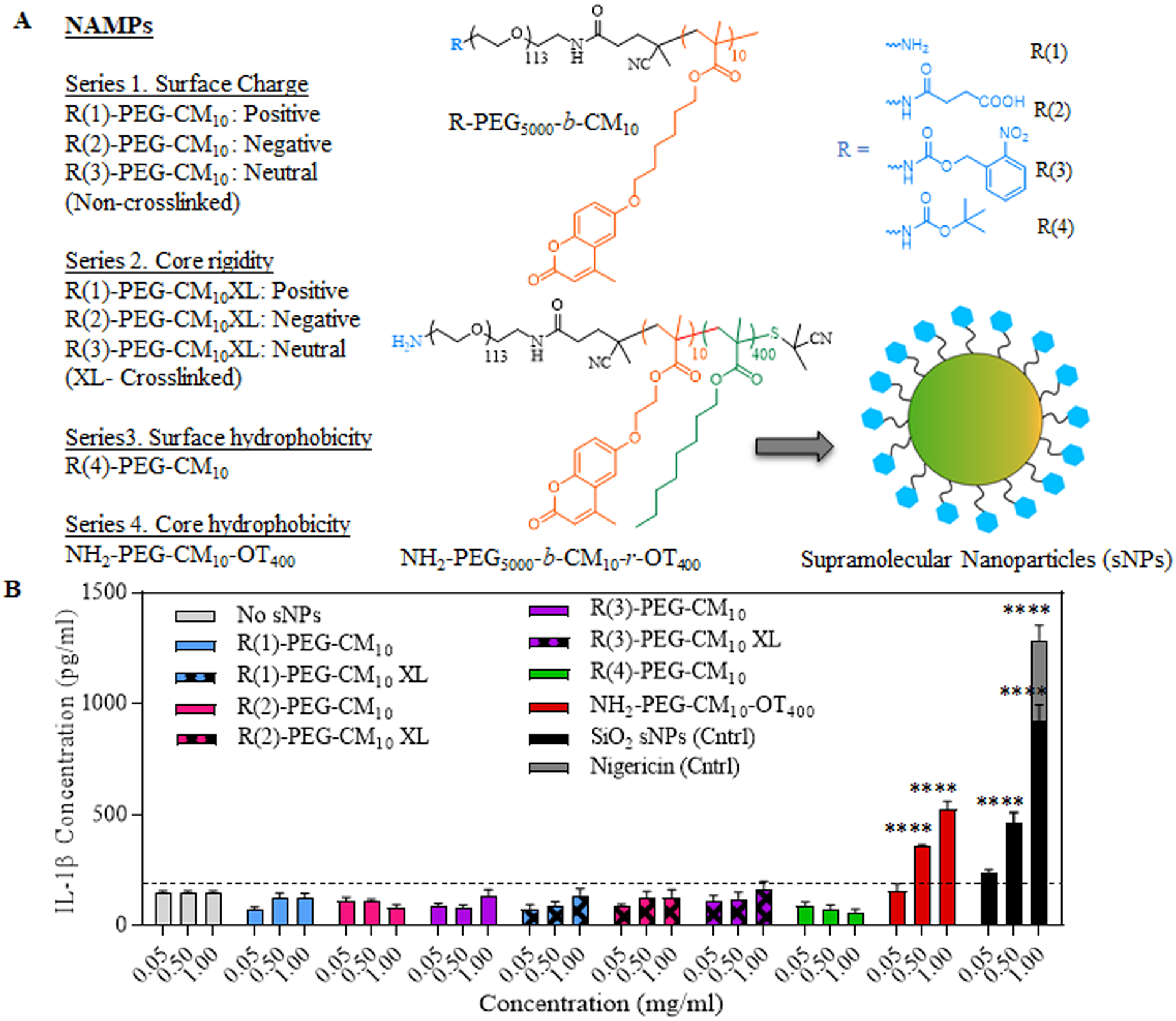
(A) Structures of different polymer constructs used to synthesize sNPs for testing varied NAMPs. Four different NAMPs tested were: Surface charge, Core rigidity, Surface hydrophobicity, and Core hydrophobicity. (B) Concentration-dependent quantification of IL-1β release by LPS-primed iBMDMs treated with a series of supramolecular nanoparticles (sNPs) containing different NAMPs as listed before. Data shown are mean ± S.E.M. (n=6). Statistical significance was determined using Ordinary one-way ANOVA and Dunnett’s multiple comparisons test. *p < 0.05, **p<0.01, ***p<0.001, ****p<0.0001.
For the preparation of different sNPs, their respective polymers were first dissolved in acetone and then stirred continuously overnight after adding 1–2 mL of Milli-Q water in a dropwise manner. (details about each series indicated in section 2 Supplementary Materials) These block copolymers being amphiphilic in nature assembles such that the hydrophobic moiety (Cum/CM-coumarin bearing alkylmethacrylate and Oct/OT-Octyl methacrylate) forms the nanoparticle core whereas the hydrophilic moiety (PEG and R) are organized on its surface (Supplementary Figure S1a).(55–58) The resultant sNP solution was further subjected to different treatments to prepare samples for cell-based assays, as shown in the supporting information. The size and ζ potential of all the sNPs are listed in the supplementary figures S2 and S3, in addition to their morphology data displayed by TEM images in figure S1b. Stability studies for core hydrophobic series sNPs exhibit stability for about a month during storage conditions (Supplementary Figure S4). Further, these four different series of sNPs denoting various NAMPs: surface charge, core rigidity, surface hydrophobicity and core hydrophobicity were examined for inflammasome activation using cell-based assays as described below.
Screening of different sNPs for their ability to stimulate LPS-primed iBMDMs for IL-1β release.
To screen the sNPs for inflammasome activation and investigate their underlying mechanisms, we used immortalized bone-marrow-derived macrophages (iBMDMs) as in vitro models.(27, 59, 60) These cells were subjected to both signals 1 and 2 (Supplementary Figure S5a) in order to test for inflammasome activation. For signal 1, the iBMDMs were primed using 50 ng/mL TLR-4 grade LPS for 12–16 hours, as previously reported by Zhong et. al. (27). For signal 2, primed cells were treated with multiple series of sNPs for a time of 24 or 48 hours (Figure 2A) with concentration variations ranging from 0.05 mg/mL to 1 mg/mL. For positive controls, the primed iBMDMs were treated with either nigericin or silicon dioxide nanoparticles, which are well known as NLRP3 activators. (20, 61, 62) The treated cells were further analyzed for IL-1β release using specific enzyme-linked immunosorbent assay (ELISA) in a dose-dependent and time-dependent manner (24 hours study: Figure 2B, 48 hours study: Supplementary Figure S5b). Recent studies have shown surface characteristics (especially charge or functionalization) as an important determinant for inflammasome activation in different nanoparticles.(31, 33, 36, 63) So, the series 1 of sNPs for IL1β screening contained varying surface charge, represented by R(1)-PEG5000-b-CM10, R(2)-PEG5000-b-CM10 and R(3)-PEG5000-b-CM10, each displaying +20, −5 and −17 surface charge, respectively (Figure 2A, Supplementary Figures S2b, S2c). Figure 2(B) and Supplementary Figure S5b show no significant IL-1β release by the cells treated with the first series of sNPs even after 48 hours of treatment, demonstrating that change in only surface charge is not sufficient to modulate the IL-1β release patterns in LPS-primed iBMDMs. We hypothesized that with uncrosslinked sNPs, the single polymer chain which is in equilibrium with its nanoaggregate to form the sNP could be the primary influence on inflammasome activation. To test this possibility, we designed crosslinked sNPs (series 2) which precludes this equilibrium. We then tested this series of the crosslinked particles, represented by R-PEG5000-b-CM10XL, on primed iBMDMs to examine their efficiency for triggering IL-1β release. Surprisingly, these particles also failed to stimulate IL-1β release from primed cells at both time points (Figure 2A, Supplementary Figure S5b), which showed that surface charge variations are indeed insufficient to stimulate and regulate inflammasome assembly even when accompanied with a rigid core. One reason for the lack of inflammasome activation could be the presence of higher PEG moieties, as reported previously in the case of silica nanoparticles, where PEGylated surface diminishes the immune response, especially NLRP3 activation.(64) Our next batch of sNPs, series 3, included a different surface modification, R(4) (Figure 2B), to test the role of nanoparticle surface hydrophobicity in inflammasome activation. This was chosen due to its role in generating immune response via cytokine production like IFN-γ and TNF-α, which pointed towards their potential for activating inflammasome (45, 65). To our surprise, this also did not show any significant increase in IL-1β release by the treated cells. As none of the surface parameters showed any influence on inflammasome activation, we next examined the nanoparticle core hydrophobicity due to the existing evidence for its association with endo-lysosomal rupture and membrane permeabilization(38, 44, 48, 49) that can serve as signal 2. For introducing hydrophobicity to the nanoparticle core, NH2-PEG-CM10-OT400 polymer was used to formulate respective sNPs. As described earlier, NH2-PEG-CM10-OT400 contains an additional 400 units of octyl hydrophobic moiety (Oct, OT400), adding hydrophobicity to the nanoparticle core (Figure 2A). sNPs obtained from NH2-PEG-CM10-OT400 polymer induced around 3.5-fold higher IL-1β release at a concentration of 1 mg/mL by primed iBMDMs, implying their potential for inflammasome activation. Different concentrations study also displayed a 2.5-fold increase in IL-1β release with sNPs amount as low as 0.5 mg/mL (Figure 2B). These findings indicate that only surface properties of the polymeric supramolecular nanoparticles, including charge variation and hydrophobicity, are not sufficient to activate inflammasome even after introducing a rigid core. In contrast, nanoparticle core hydrophobicity plays a pivotal role in stimulating IL-1β release in LPS-primed iBMDMs.
Correlation of differential nanoparticle core hydrophobicity with IL-1β release indicating inflammasome activation.
After determining core hydrophobicity as a potent IL-1β stimulator, we investigated if systematic variations in the core hydrophobicity of sNPs influence or correlate with inflammasome activation. To check this, we synthesized hydrophobic series-4 sNPs with different y values (y represents the repeating units of octyl hydrophobic moiety “OT”), varying from 0 to 400, with ‘0’ as no hydrophobicity and ‘400’ as highest (Figure 3A), as mentioned earlier. LPS-primed iBMDMs were then treated with three different concentrations (1, 0.5, 0.05 mg/mL) of hydrophobic series sNPs (y=0 to y=400) for 24 and 48 hours. This was followed by quantification of IL-1β released in the cell supernatant using ELISA along with the estimation of viable cells using MTT assay (Figure 3B, Supplementary Figure S6). Figure 3B displays a remarkably higher IL-1β secretion by the cells treated with hydrophobic series sNPs starting from ‘y’ as low as 100 units than just LPS-primed cells, both in 1 mg/mL and 0.5 mg/mL concentrations study. However, there is no significant difference in IL-1β release when the cells are treated with sNPs, completely lacking the octyl units (y=0) with respect to untreated primed cells. To evaluate if these treated cells are also undergoing cell death along with IL-1β production (known as pyroptosis, last stage of inflammasome activation(2)), we measured the percent viability of primed cells after 24 hours of sNPs (hydrophobic series, y=0 to y=400) treatment. As shown in figure S4, there is a significant reduction in percent viability, directing towards increased cell death mediated by hydrophobic particles starting from y=100 to y=400 compared to only LPS-primed cells. Moreover, there is no significant difference in percent viability of y=0 sNPs treated cells with respect to only primed cells, which is consistent with our findings in figure 3B, proving no inflammasome activation by the less hydrophobic particles. Altogether, the above findings suggest an important role of nanoparticle core hydrophobicity in activating inflammasome and point it out as a potential inflammasome-activating NAMP.
Figure 3. Correlation of nanoparticle core hydrophobicity with inflammasome assembly and activation.
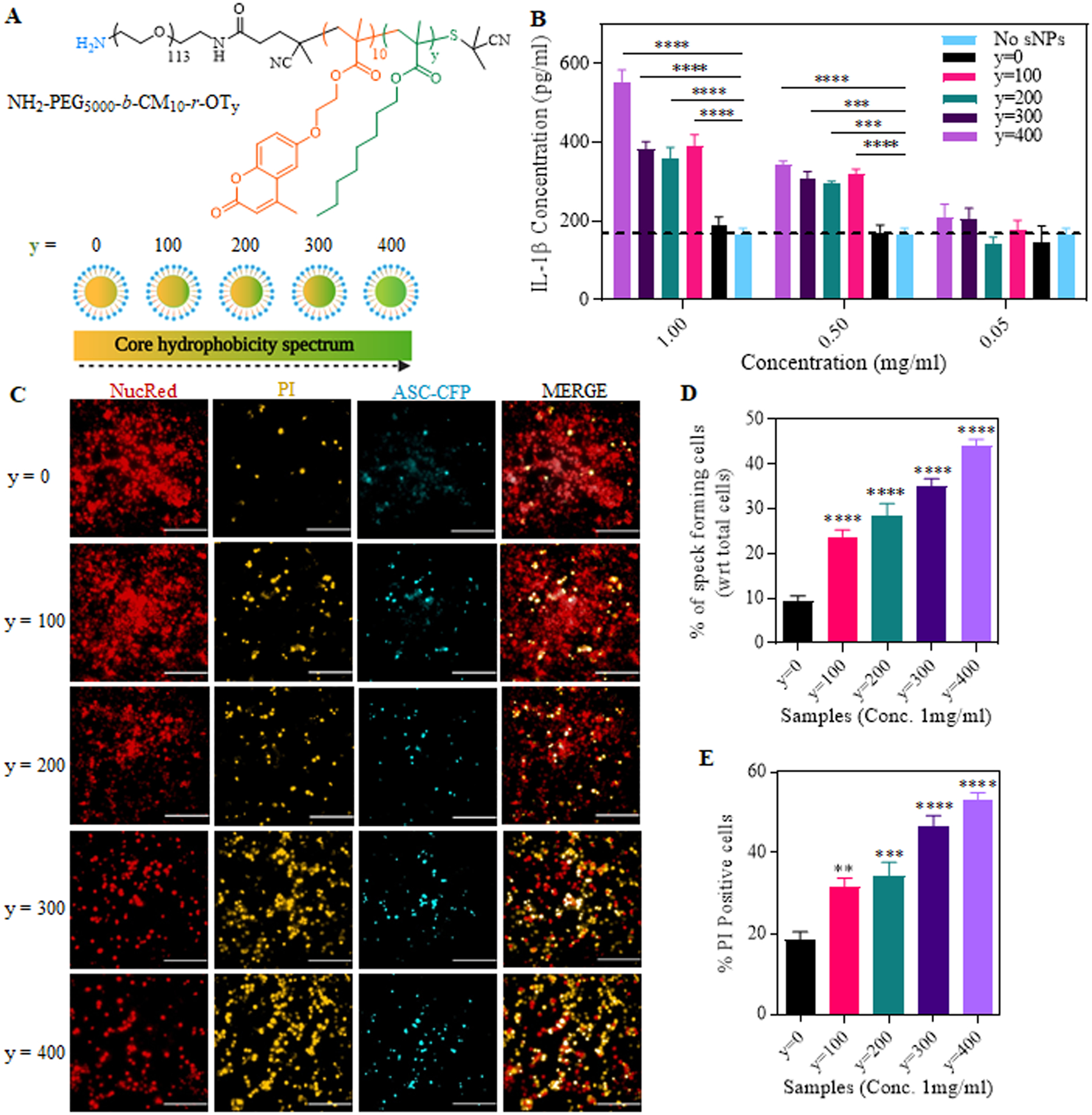
(A) Structure of sNPs containing sequentially varied core hydrophobicity due to different octyl moieties represented by “y”, ranging from y=0 to y=400. (B) IL-1β release in the supernatant of LPS-primed iBMDMs treated with sNPs with sequentially varied core hydrophobicity. Data shown are mean ± S.E.M. (n=6). Statistical significance was determined using Ordinary two-way ANOVA and Dunnett’s multiple comparisons test. *p < 0.05, **p<0.01, ***p < 0.001, ****p<0.0001. (C) Representative High Content Analysis (HCA) microscopic images of NucRed and PI stained iBMDMs, already primed and treated with y=0 to y=400 sNPs. Red fluorescence denotes NucRed signal, CFP channel shown in blue indicates the ASC specks, PI signal is shown by TRITC channel shown in yellow. Scale bars represent 200 μm. Images were taken at 20x magnification. (D) Percentage of speck forming cells after 16 hours of sNPs treatment, normalized to the total number of live cells counted by NucRed signal. (E) Graph shows the percentage of dead iBMDMs after 16 hours of sNPs treatment, indicating percent pyroptosis. This was again normalized with the total signal of live (NucRed) and dead (PI) cells. Data shown are mean ± S.E.M. (n=6). Statistical significance was determined using Ordinary one-way ANOVA and Dunnett’s multiple comparisons test. *p < 0.05, **p<0.01, ***p < 0.001, ****p<0.0001.
Nanoparticle core hydrophobicity directly correlates with NLRP3 inflammasome complex assembly and activation.
We next hypothesized that hydrophobic sNPs IL-1β response is due to NLRP3 type inflammasome assembly leading to caspase-1 activation. To examine this, we treated the cells that were knockout for either NLRP3 or caspase-1, with 1 mg/mL hydrophobic series sNPs (y=0 and y=400) and quantified IL-1β release in the cell supernatant. We compared this response with that of WT iBMDMs and found that the IL-1β activation and release is significantly abrogated in NLRP3 as well as caspase-1 deficient iBMDMs, pointing towards the involvement of these proteins in this signaling response (Supplementary figure S7). Having established the involvement of NLRP3 and caspase-1 proteins, we next evaluated if variation in nanoparticle core hydrophobicity correlates with their activation. To further gain mechanistic insights, we investigated the effect of nanoparticle core hydrophobicity on the expression of various NLRP3 inflammasome-associated proteins. We visualized and quantified the adaptor protein, ASC (apoptosis-associated speck-like protein containing a CARD) speck formation in treated cells over time to analyze the inflammasome complex assembling with varying core hydrophobicity. For this, we chose to utilize iBMDMs stably expressing CFP tagged ASC protein for speck visualization. ASC specks are a common indicator of inflammasome assembly that can be visualized using a confocal microscope when tagged with a fluorescent marker. (66–68) ASC usually remains dispersed in the cytosolic region of a cell; however, the assembly of inflammasome complex recruits all the ASC adaptor molecules to bind the threat sensor and effector enzyme together. This results in a very bright detectable punctated ASC speck. As NLRP3 complex assembly and activation are a binary all-or-none phenomenon (69), there is only one speck per inflammasome activated cell, thus providing a clear distinction for imaging and visualization of inflammasome complex assembly in a cell population.
To attain this, ASC-CFP iBMDMs were first primed with TLR4-grade LPS, then treated with the core hydrophobic series of sNPs (y = 0 to 400). Finally, these treated cells were incubated with NucRed Live 647 Ready probes and propidium iodide (PI) for staining live cell nuclei and dead cells, respectively, and imaged with a High Content Analysis (HCA) microscope. Fluorescence microscopic images display a significantly decreasing NucRed and increasing PI signal with higher core hydrophobicity of sNPs than y=0 sNPs at 16 hours time point, exhibiting a positive correlation with the cell death. Moreover, microscopic images and analysis reveal a significant increase in ASC punctated specks (shown in CFP channel) in y=100 to y=400 sNPs treated cells with respect to y=0, which shows no or low number (Figure 3C and 3D). Figure 3C displays ASC punctated bright fluorescent dots in y=100 to 400 treatment groups in the CFP channel, clearly indicating inflammasome complex assembly. We also observed a positive correlation of ASC speck forming cells and nanoparticle core hydrophobicity, as displayed in Figure 3D. As mentioned before, the dead cells were stained with PI to determine percent pyroptosis, represented by percent PI-positive cells in Figure 3E. The graph shows a significantly higher number of PI-positive cells with increasing nanoparticle core hydrophobicity and a direct correlation similar to ASC speck formation. Altogether from microscopy data, it is evident that inflammasome complex assembly and pyroptosis are positively correlated with nanoparticle core hydrophobicity. We next wanted to look at the expression levels of various NLRP3 inflammasome signaling associated proteins (NLRP3, ASC and pro-caspase-1) in the lysate of treated cells using western blotting (Figure 4A, iii). We also examined and quantified the expression of active caspase-1 released in the supernatant of treated cells (Figure 4A, iv) to confirm if nanoparticle core hydrophobicity also modulates caspase-1 activation and has any correlation similar to complex assembling. As exhibited in figure 4B, protein expression of all the inflammasome assembly components, NLRP3, ASC, and pro-caspase-1, is consistent in all the treatment groups. Simultaneously, there is a significantly higher release of active caspase-1 in the cellular supernatant obtained after treatment with increasing hydrophobicity of sNPs (Figure 4B). Statistical analysis in figure 4C also shows a trend in caspase-1 activation with higher nanoparticle hydrophobicity, pointing towards a direct correlation among them during NLRP3 inflammasome signaling. Overall, increasing core hydrophobicity exhibits a greater number of specks forming cells, PI-positive cells, and higher expression of active caspase-1 in the supernatant. These observations indicate a positive correlation of nanoparticle core hydrophobicity with that of NLRP3 assembly and other downstream processes, including caspase-1 activation and pyroptosis.
Figure 4. Expression of the NLRP3-related proteins in the sNPs treated iBMDMs.
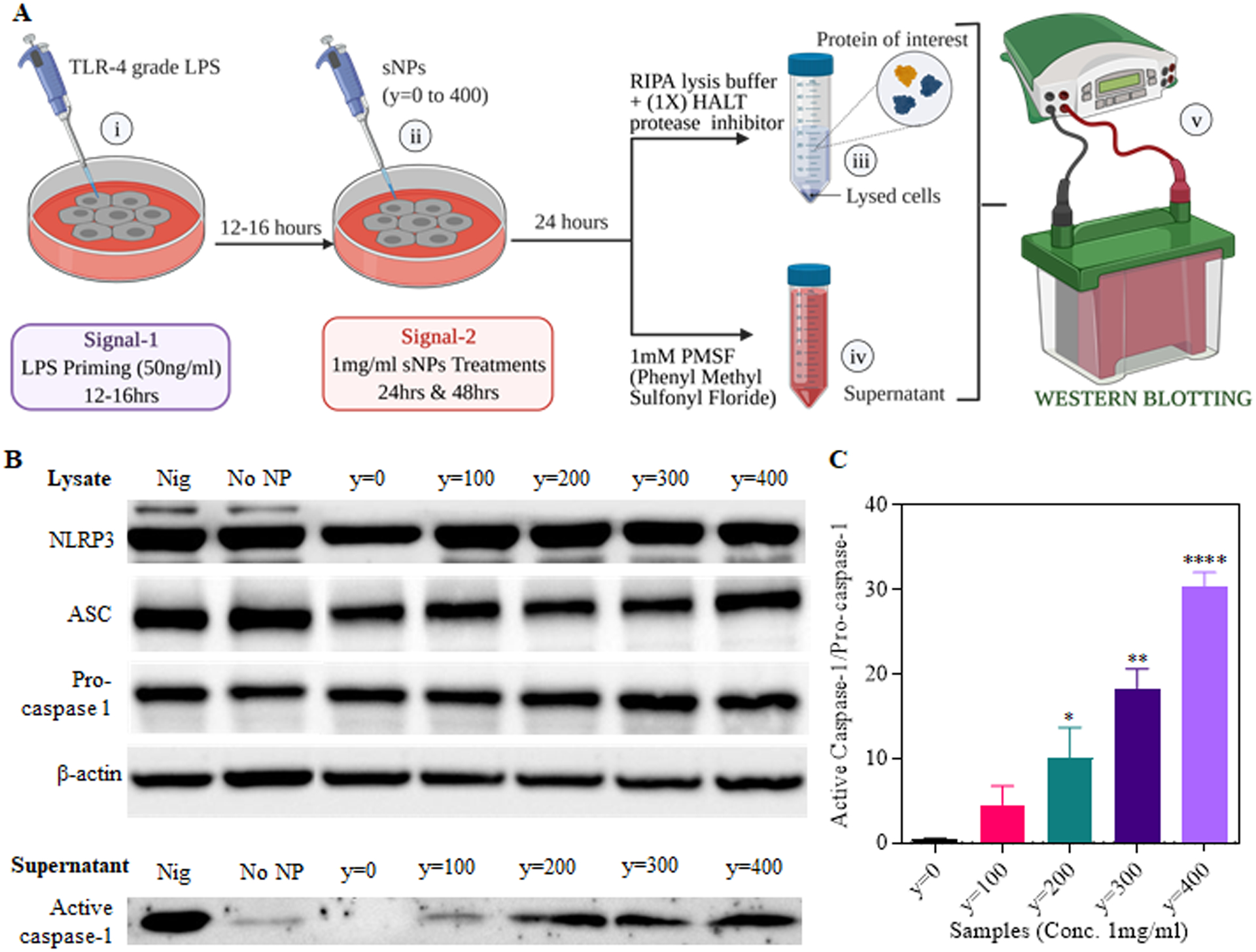
(A) Outline of treatments and sample processing for analysis of protein expression levels after inflammasome activation using western blot assay. i) Signal 1, LPS priming of iBMDMs for 12–16 hours. ii) Signal 2, y=0 to 400 sNPs treatment for 24hours. iii) Protein lysate preparation using RIPA buffer with protease inhibitor. iv) Supernatant collection followed by the addition of 1mM PMSF before storage. v) Western blot analysis of NLRP3-related proteins in the sample lysate as well as supernatant. (B) Top part shows the protein expression levels of NLRP3, ASC, Pro-caspase-1 determined by western blot of cell lysate obtained from sNPs treated iBMDMs. Bottom part displays the protein levels of active caspase-1 subunit (p20) released in the supernatant of these sNPs treated iBMDMs. No NP group represents no nanoparticle treatment. (C) Graph displaying the quantitative analysis of active caspase-1 released in the supernatant, normalized to pro-caspase-1 protein expression in the cell lysate. Data shown here are mean ± S.E.M. (n=3). Statistical significance was calculated by Ordinary one-way ANOVA and Dunnett’s multiple comparisons test. ‘ns’, not significant; *p < 0.05, **p<0.01, ***p < 0.001, ****p<0.0001.
Lysosomal rupture as the first underlying mechanism for activating NLRP3 inflammasome by nanoparticle core hydrophobicity.
We were interested in investigating the underlying signaling pathways after determining that nanoparticle core hydrophobicity is directly correlated to NLRP3 inflammasome assembly and activation. To address this, we began with testing the lysosomal rupture pathway, which is commonly associated with NLRP3 activation via cathepsin B maturation and is known to be impacted by nanoparticle hydrophobicity (35, 40, 48, 70). The lysosomal disruption mediated by series 4 sNPs was examined using both lysotracker and acridine orange lysosomal staining dyes in independent experiments. To evaluate the change in lysosomal rupture due to varying core hydrophobicity, we first determined the cellular uptake efficiency of these sNPs by making them fluorescent and calculating their internalization using flow cytometry and microscopy. To synthesize fluorescent y=0 to y=400 sNPs, DiD (1,1-Dioctadecyl-3,3,3,3-tetramethylinodicarbocyanine) labeling dye was encapsulated in different sNPs of hydrophobic core series. To test the stability of these DiD-loaded sNPs, we evaluated the free dye leaching out of the particles over a time course of 7 days by quantifying the absorbance for dye encapsulated particles at 660nm. Supplementary Figure S8 shows that the absorption spectra of DiD-loaded nanoparticles remains unchanged even after 7 days suggesting that there is no leakage of dye from these particles. After the synthesis of stable DiD encapsulated sNPs, the cells were labeled with CFSE (Carboxyfluorescein succinimidyl ester) followed by LPS treatment to obtain CFSE-positive primed iBMDMs. These CFSE stained iBMDMs were further incubated with fluorescent hydrophobic series nanoparticles (y=0 to y=400) for indicated time points, and double-positive cells were evaluated using flow cytometry. As seen in Figure 5A, 100 percent of the CFSE positive cells internalize fluorescent nanoparticles at 37°C starting from 3hours itself and are consistent even in 9 hours, showing that internalization is consistent in all the treatment groups. Moreover, this response is significantly inhibited upon treatment at 4 °C, suggesting that the uptake is via one of the internalization modes and not just passive dye diffusion. The cells were then stained with Lysotracker Red DND 99 after DiD sNPs (y=0 to 400) treatments and imaged using confocal microscopy for quantifying intact lysosomes. As observed in Figure 5B, even though DiD channel (red) shows a similar signal in all the treatment groups, the lysotracker signal drastically reduces with increasing core hydrophobicity of nanoparticles. Statistical quantitation of these microscopic images is shown in Figure 5C, which exhibits the mean fluorescence percent of lysotracker (TRITC channel) in sNPs treated cells with respect to LPS primed cells. We observed a significantly lower percentage of lysotracker signal in y=300 and y=400 sNPs treated cells than y=0 ones. Consistent with this finding, we also observed a significant decrease in orange signal (Supplementary Figures S9a, S9c) and an increase in green signal using flow cytometry (Supplementary Figures S9b, S9d) in acridine orange stained iBMDMs at 4hours treatment, pointing towards significantly higher lysosomal rupture in y=400 treatment group. This corroborated with earlier observation and thus indicated a heightened lysosomal disruption with increased particle hydrophobicity. Thus, these results suggest that particles falling in the highest spectrum of core hydrophobicity induce inflammasome activation via the lysosomal disruption pathway. We next investigated if mature cathepsin B is also released in these cells with maximum lysosomal disruption, as it is previously known to act as the connecting element between lysosomal rupture and inflammasome activation (19, 36, 71). As shown in Figure 5D, we observed a notably higher expression of mature cathepsin B in cell lysate treated with more hydrophobic sNPs. There is also a sequential trend in the increased activation of the enzyme with changing hydrophobicity (Figure 5E) when normalized with β-actin. These observations collectively indicate that high core hydrophobicity leads to lysosomal disruption, which activates cathepsin B resulting in NLRP3 activation. Moreover, nanoparticle core hydrophobicity positively correlates with the maturation of cathepsin B leading to varying degrees of inflammasome activation.
Figure 5. Cellular uptake of DiD encapsulated fluorescent sNPs and their lysosomal rupture potential in treated iBMDMs.
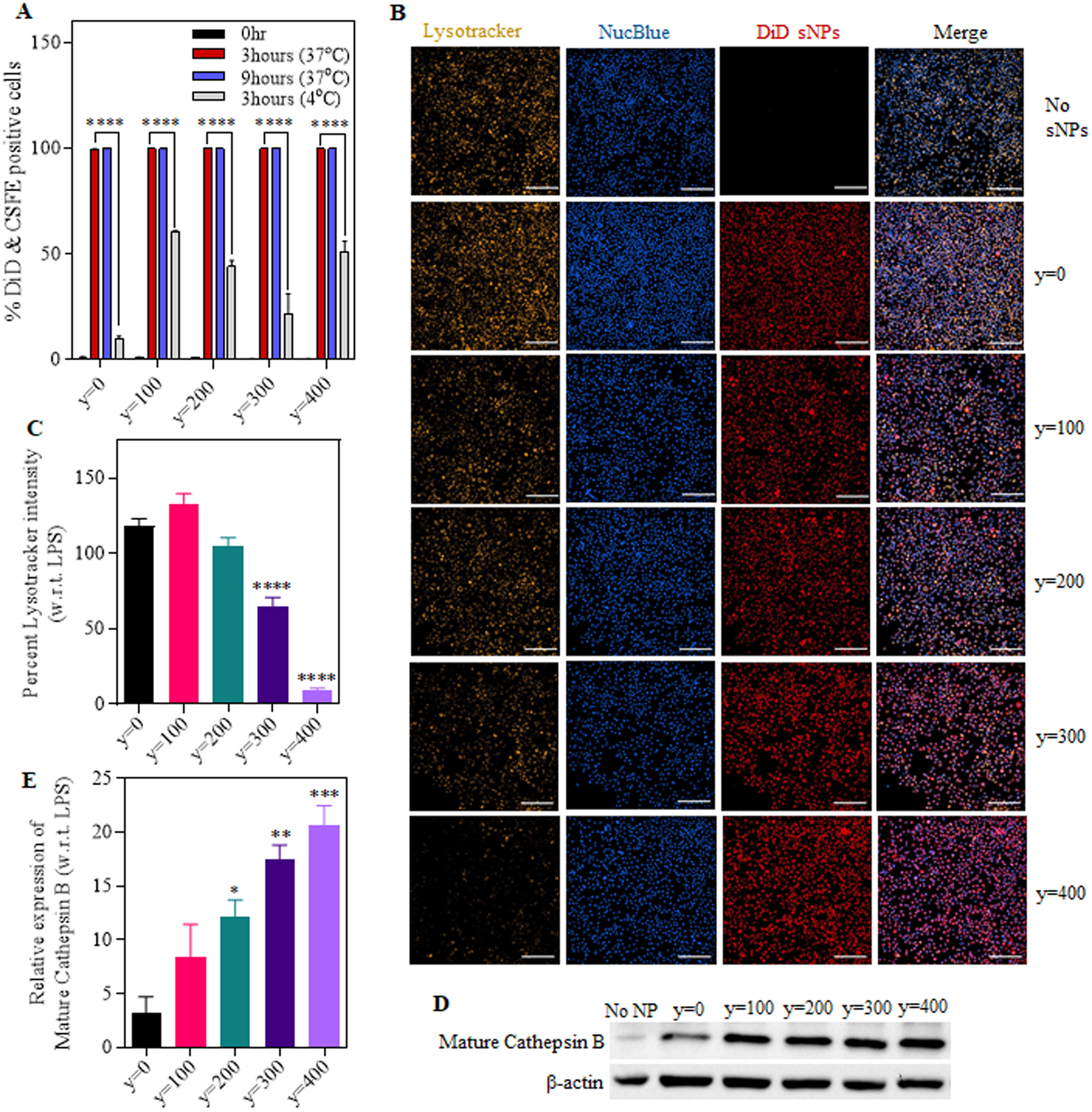
(A) Graph shows the percentage of CSFE labeled cells internalizing DiD dye encapsulated sNPs at different time points (0, 3 and 9 hours). (B) Fluorescent images of iBMDMs treated with DiD sNPs (y=0 to y=400) for 4hours, followed by lysosomal labeling with Lysotracker Red DND-99 and nuclear staining with NucBlue Live ReadyProbes Reagent (Magnification: 20x, Scale bar: 200 μm). (C) Quantification of percentage lysotracker mean fluorescence intensity in sNPs treated iBMDMs with respect to primed iBMDMs. (D) Representative western blot image showing the expression of mature cathepsin B in treated and only LPS primed iBMDMs. (E) Graph displaying a relative expression of mature cathepsin B in sNPs treated iBMDMs compared to only primed ones; normalized with b-actin expression in the respective samples. Data shown here are mean ± S.E.M. (n=3). Statistical significance for (A) was calculated using two-way ANOVA and Dunnett’s multiple comparisons test. Statistical significance for (C) and (E) were calculated by Ordinary one-way ANOVA and Dunnett’s multiple comparisons test. ‘ns’, not significant; *p < 0.05, **p<0.01, ***p < 0.001, ****p<0.0001.
Calcium influx followed by mtROS production act as another underlying mechanism for activating NLRP3 inflammasome by nanoparticle core hydrophobicity.
Another underlying signaling known to be associated with NAMP activating NLRP3 inflammasome is the Ca2+ influx – mitochondrial ROS production pathway. (27, 72) To examine whether these nanoparticles also follow this pathway in addition to the lysosomal disruption signaling, we first assessed calcium flux in iBMDMs treated with y=0 to y=400 series of sNPs as compared to only LPS-primed iBMDMs. To measure intracellular Ca2+ levels, we stained the treated cells with a cell-permeant calcium indicator, Fluo-4AM, and analyzed them using either flow cytometry or confocal microscopy (Supplementary Figure S10a). Microscopy data in Figure 6A revealed a significantly higher calcium influx in y=400 sNPs treated cells as compared to y=0, suggesting its critical role in NAMP mediated inflammasome activation. Moreover, (Supplementary Figures S11a and 11c) provide evidence for significant inhibition upon treatment with BAPTA-AM chelator. Next, we determined the fold change in median fluorescence intensity of sNPs treated primed cells compared to only primed cells at indicated time points (4, 12, and 24 hours) using flow cytometry, as displayed in Figure 6B. The graph indicates a remarkably higher intracellular calcium in the cells treated with y=100, 300, and 400 sNPs at 12 hours. At 24 hours, it shows a significantly higher calcium flux even in y=200 treated group, posing that these different hydrophobic nanoparticles exhibit different time-dependent trends of calcium influx. Further, to test if this response is inhibited by the use of calcium chelator, we incubated the y=400 sNPs 12 hours treatment group with BAPTA-AM (as it showed maximum signal) and found Fluo-4 signal to be significantly inhibited, Supplementary Figure S10b. Previous reports have also shown a clear association of calcium influx with multiple organelle damage, especially mitochondrial damage.(32, 73, 74) So, we tested mitochondrial ROS production previously shown to activate the NLRP3 inflammasome directly. (75) Supplementary Figure S10a also illustrates the schematics for different treatments followed by MitoSOX staining to calculate the mitochondrial ROS production at different time points. Again, we performed a time-dependent study using flow cytometry, but a later time point study using microscopy for identifying mitochondrial ROS (mt ROS) production as it occurs downstream to calcium influx. We noticed a significantly higher mitochondrial ROS production in the cells treated with y=400 sNPs after 16 hours in microscopy (Figure 6C), which gets significantly inhibited upon treatment with a calcium chelator suggesting that most of the mt ROS production is as a result of calcium flux leading to mitochondrial damage (Supplementary Figure S11b and S11d) Figure 6C and Supplementary Figure S11b represents MitoSOX, and NucBlue stained cells treated with hydrophobic core series sNPs, and Supplementary Figure S11d depicts its statistical analysis as relative fluorescence of treated groups to untreated, only primed ones. Flow cytometry analysis also shows a significant fold increase in median fluorescence intensity of y=400 treatment group at 24 hours time point in compliance with the microscopy data (Figure 6D), which significantly inhibits upon BAPTA treatments (Supplementary Figure S10c). Additionally, y=300 sNPs treated cells also display a remarkable fold change at 24 hours. These results demonstrate that polymeric supramolecular nanoparticles varying in their core hydrophobicity follow different signaling pathways to activate the NLRP3 inflammasome. Altogether, these findings demonstrate that particles with varying hydrophobic core show a different trend and kinetics for calcium influx leading to mt ROS production, indicating an association of particles hydrophobicity with distinctive calcium influx patterns, causing different degrees of inflammasome activation.
Figure 6. Intracellular Ca2+ levels and mitochondrial ROS production in the sNPs treated primed-iBMDMs.
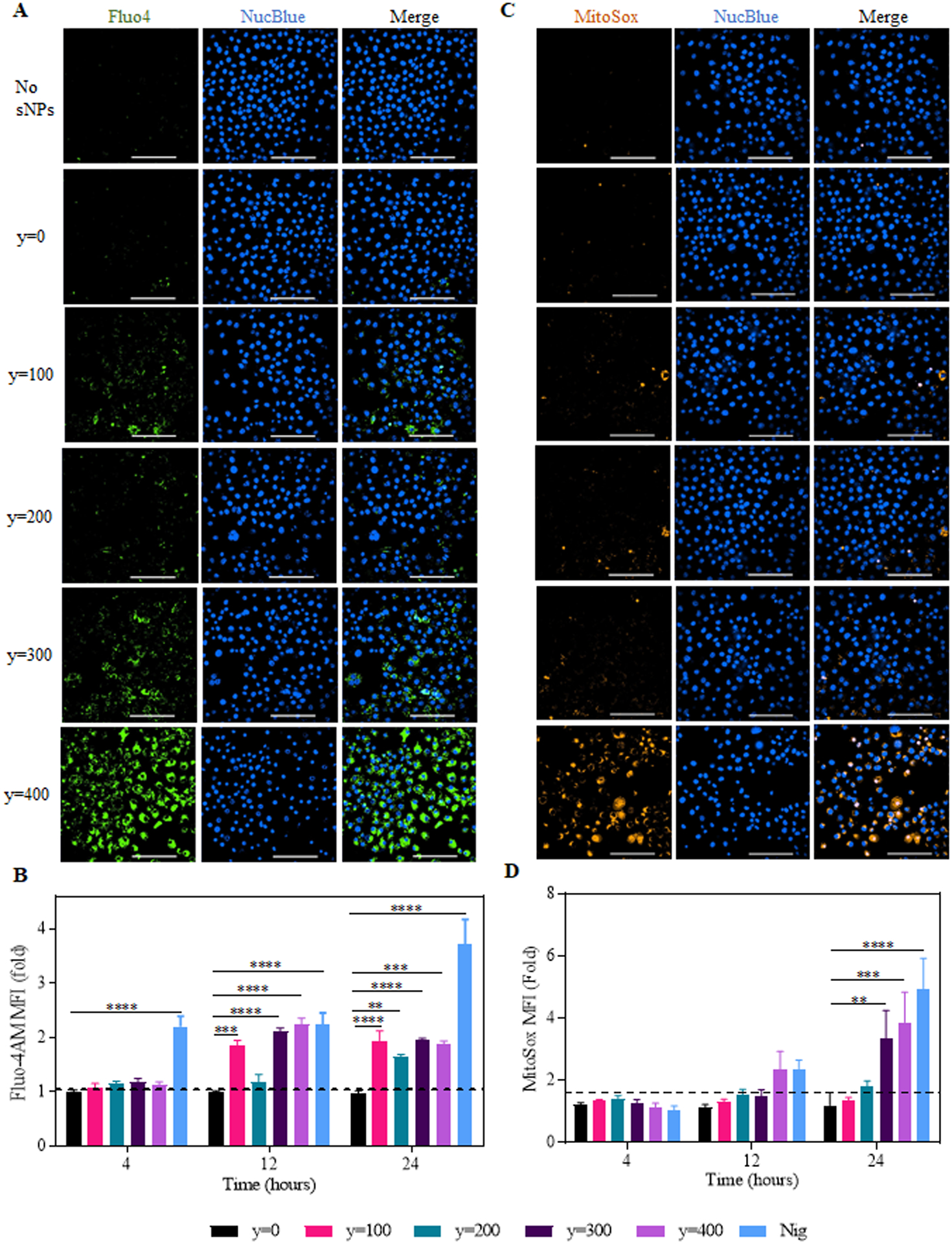
(A) Representative microscopic images of fluo-4AM stained iBMDMs treated with sNPs. Nuclei were stained with NucBlue Live ReadyProbes Reagent. Green shows intracellular calcium, and blue denotes the live-cell nucleus. Images are of 60x magnification (Scale bar: 100 μm). (B) Graph displays time-dependent fold change in the median fluorescence intensity of fluo4 in sNPs (y=0 to 400)-treated cells compared to untreated. Data shown here are mean ± S.E.M. (n=3). Statistical significance was calculated by Ordinary one-way ANOVA and Dunnett’s multiple comparisons test. ‘ns’, not significant; *p < 0.05, **p<0.01, ***p<0.001, ****p<0.0001. (C) Representative confocal images of iBMDMs treated with y=0 to y=400 sNPs for 16hours, followed by MitoSOX and NucBlue staining. MitoSOX (Orange) represents mitochondrial ROS, and NucBlue (Blue) shows live-cell nucleus. Images are of 60x magnification (Scale bar: 100 μm). (D) Graph showing fold change in median fluorescence intensity of MitoSOX stained treated cells with respect to untreated at indicated time points. Data shown here are mean ± S.E.M. (n=6). Statistical significance was calculated by two-way ANOVA and Dunnett’s multiple comparisons test. *p < 0.05, **p<0.01, ***p < 0.001, ****p<0.0001.
DISCUSSION
Polymeric nanoparticles have gained increasing attention for immunotherapy applications, including the delivery of immunomodulators due to their modularity and scalability over other nanoplatforms. However, there has not been any systematic, comprehensive study to understand the effect of various polymeric attributes on immune responses like inflammasome activation. NAMPs represent a novel class of molecular signatures in addition to conventionally known PAMPs and DAMPs, which interact with and activate the innate immune NLRP3 sensor. (16, 30, 34) Most previous studies have listed inflammasome-activating biomaterials; still, only some have attempted to identify the NAMPs (from different types of nanoparticles) leading to NLRP3 assembly(31–33, 35–43), and among them, very few focused on polymeric nanosystems(23, 36–38). In the present study, we have employed a multiparametric designer supramolecular polymeric nano-platform to systematically analyze the influence of several distinctive features of NAMPs on NLRP3 assembly and activation in a single polymeric system. Moreover, several cell-based assays helped identify the underlying mechanisms potentially leading to inflammasome activation to provide a complete picture. This is the first study that has provided a designer platform for systematically varying both surface and core properties in the same polymeric nano-system one at a time and comprehensively evaluate their influence on inflammasome activation. Here, we utilize four series of these designer polymeric sNPs, each containing one tweaked NAMP, represented as surface charge, surface hydrophobicity, core rigidity or core hydrophobicity. These sNPs were synthesized from a class of block co-polymers with multiple flexible moieties: “R”, “Cum” and “Oct”, that help in introducing surface variation (charge and hydrophobicity), core rigidity (cross-linkage), and core hydrophobicity (sequential variation), respectively. The block-co-polymer design consists of polyethylene glycol (PEG) moieties that provide hydrophilicity to the structure, octyl methacrylate (Oct) moieties that provide core hydrophobicity to the structure, coumarin methacrylate (Cum) moieties that provide core rigidity via crosslinking the structure and R functional groups that provide variations in surface properties of sNPs.
Here, we first examined nanoparticle surface properties as they serve as the initial contact points during cellular interaction and uptake. Surprisingly, contrary to other nano-systems (31, 36, 43), our platform does not activate NLRP3 inflammasome upon varying surface charge or hydrophobicity, leading us to conclude that surface variation is insufficient for eliciting NLRP3 activation in these polymeric nanoparticles. This response is similar when these particles are UV-crosslinked to attain a rigid core. One of its reasons could be the presence of numerous PEG repeating units as the first block in these polymeric sNPs, which could diminish inflammasome activation similar to PEGylated silica particles’ activity previously reported by Marzaioli et al. (63) Besides, for the first time, our study reveals core hydrophobicity as a novel polymeric NAMP that can regulate NLRP3 activation, even in the presence of multiple PEG units. This highlights the importance of considering core hydrophobicity in addition to surface functionalization and PEGylation while developing novel polymeric delivery systems for various immunotherapeutics and immunodiagnostic applications. In addition to identifying core hydrophobicity as a novel NAMP, this study also discovered its positive correlation with ASC speck formation, caspase 1 activation and pyroptosis, indicating its direct relationship with NLRP3 assembly and activation. This provides us a structure-based polymeric activator that can be modulated to attain varying degrees of NLRP3 activation to design a chemically controlled system advantageous for vaccine development and immunotherapy.
Furthermore, mechanistic studies exhibited the role and interplay of multiple signaling cascades towards the activation of NLRP3 inflammasome mediated by nanoparticle core hydrophobicity. Here, we found that both lysosomal-rupture mediated cathepsin-B release and mitochondrial-ROS production via calcium influx pathways strive together to generate varying levels of inflammasome activation. Our study displays remarkably increased lysosomal rupture and cathepsin B maturation by nanoparticles with the highest core hydrophobicity. One explanation involves the presence of numerous octyl repeat units “OT,” possibly causing the membrane destabilization in a lysosomal environment. This observation is similar to previous studies where butyl, octyl, and other hydrophobic repeat units in different block co-polymeric nano-systems mediate endo-lysosomal rupture and escape. (38, 48, 76–78) Moreover, consistent with other studies, we also observed an increase in cathepsin B maturation followed by lysosomal rupture, a well-known NLRP3 activator.(70) Additionally, we obtained a sequential increase in cathepsin B maturation with varying core hydrophobicity, again pointing towards a positive correlation that is advantageous for the system’s modular capacity. Besides the lysosomal disruption pathway, we also reported calcium flux followed by mitochondrial ROS production as a secondary pathway for mediating NLRP3 activation by these hydrophobic core sNPs. We observed that higher core hydrophobicity leads to increased calcium influx resulting in greater mtROS production. This could result from plasma membrane lipid peroxidation leading to calcium influx and mtROS production similar to other nanosystems. (73) Generation of mtROS, in turn, has been clearly shown to stimulate NLRP3 assembly in previous studies. (75)
Overall, this comprehensive mechanistic study reveals that differential nanoparticle core results in varying degrees of inflammasome activation by following distinctive trends, degrees, and kinetics of two underlying signaling pathways: lysosomal disruption – cathepsin B maturation and calcium influx – mt ROS production signaling cascades as described further. The most hydrophobic group, y=400, follows both the pathways early on, whereas y=300 displays lysosomal disruption as its prominent pathway to activate inflammasome as shown by lysotracker staining and mature cathepsin B expression results. Moreover, y=100 and y=200 sNPs, mediate some degree of NLRP3 activation due to both pathways but favorably disposed towards one. In such cases (y=100/200), only one cellular damage process is not sufficient to activate NLRP3 to a noticeable extent; multiple pathways are required. However, there is more inclination towards a single pathway that dominates the entire process. Our study indicates that y=100 sNPs exhibit a preference for calcium influx, as there is a remarkably increased signal in 12 hours. In contrast, y=200 sNPs are more directed towards cathepsin B maturation. Our findings provide evidence towards a positive correlation between NLRP3 activation and nanoparticle core hydrophobicity, the novel NAMP, which is attained by following different cellular signaling cascades. Additionally, sNPs series 1, 2 and 3 reveal that sole variation in either surface charge, surface hydrophobicity, or core rigidity is insufficient to elicit NLRP3 activation.
Taken together, this study provides a better understanding of immune cells-nanomaterials interaction, which could help in tuning the NAMPs either to obtain safer platforms for diagnostics and therapeutics; or to derive controlled inflammasome-activating platforms that can generate vaccine adjuvants (51–53, 79) and agonists for immunotherapy. We anticipate our findings to serve as a guiding principle for screening multiple other parameters and different types of nanoparticles and microparticles in order to execute inflammasome activation, thus providing a way to engineer better serving biomaterials.
MATERIALS AND METHODS
Materials / Reagents:
All the reagents were purchased from commercial suppliers (ThermoFisher, Invivogen, Tocris, Adipogen, Biolegend, Cell Signaling Technology, Biorad, Sigma-Aldrich). Ultrapure lipopolysaccharide (LPS) and Nigericin were purchased from Invivogen and Tocris, respectively. Western antibodies: anti-NLRP3/NALP3 (mouse) monoclonal antibody (Cryo-1), anti-Asc polyclonal antibody (AL177) and anti-Caspase-1(p20)(mouse) monoclonal antibody (Casper-1) was purchased from Adipogen Life Sciences, whereas anti-β-actin antibody was procured from Biolegend. Anti-rabbit IgG, HRP-linked secondary antibody was ordered from Cell Signaling Technology. For protein lysate and sample preparation, RIPA lysis buffer, Halt™ Protease and Phosphatase Inhibitor single-use cocktail EDTA-Free (100X), and Alfa Aesar Laemmli SDS sample reducing 4X buffer were procured from ThermoFisher Scientific. For western, TGX Stain-Free™ FastCast™ Acrylamide Kit, 10% and Clarity™ Western ECL substrate were purchased from Biorad. The chemicals for preparing western buffers were procured from Sigma-Aldrich, including TRIZMA base, sodium dodecyl sulfate, glycine, sodium chloride, tween- 20 bovine serum albumin (BSA), methanol and 2-mercaptoethanol. The MTT compound (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was obtained from Promega, and IL-1β mouse uncoated ELISA kits were obtained from ThermoFisher Scientific. Cell-culture media ingredients like DMEM, heat-inactivated Fetal Bovine Serum (FBS) and Penicillin-Strep were purchased from Gibco, Life Technologies. All other cell staining compounds and dyes, including Propidium iodide, NucRed™ Live 647 ReadyProbes™Reagent, NucBlue™ LiveReadyProbes™Reagent (Hoechst 33342), LysoTracker™ Red DND 99, Fluo-4AM, MitoSOX and DiD dye (DiIC18(5); 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt) were procured from ThermoFisher Scientific. All forms of immortalized bone-marrow-derived macrophages (iBMDMs) were obtained from Fitzgerald Lab, UMass Medical School, Worcester.
Methods:
Synthesis and characterization of supramolecular polymeric nanoparticles (sNPs)
The detailed synthesis and characterization of polymer construct are described in the Supplementary Materials section. For synthesizing different supramolecular polymeric nanoparticles, 2 mg of respective polymer (as indicated in Figure 2A) was dissolved in 150 μL acetone. To this solution, 1 mL of water was added in a dropwise manner and stirred continuously overnight. This results in acquiring mono-dispersed micelle polymeric nanoparticles in water. To generate rigid core crosslinked sNPs (wherever mentioned), these micelle solutions were then put into the UV incubator with a 365nm lamp for 3minutes. Afterward, all these particles were concentrated using 3k MWCO centrifugal filters and redispersed in PBS buffer to obtain a stock of 10 mg/mL for the following cell-based assays. In most assays, the working concentration was 1 mg/mL unless it was a concentration-dependent study. For synthesizing fluorescent nanoparticles, sNPs were encapsulated with DiD dye.
Size of DLS characterization:
Dynamic Light Scattering (DLS) data were recorded by a Malvern Nanozetasizer ZS90 with a 637-nm laser source with non-invasive backscattering technology detected at 173° using disposable sizing cuvette. Sample micelle and nanogel were measured at a concentration of 0.2 mg/mL.
Zeta potential of nanoparticles:
To measure the zeta potential of each nanoparticle, 1 mL nanoparticle solution at a concentration of 0.2 mg/mL was injected into Disposable folded capillary cells (DST1070) by a 1 mL syringe to avoid bubble. The data was recorded by a Malvern Nanozetasizer ZS90.
Stability of NH2-PEG5000-b-CMx-r-OTy sNPs:
For stability studies, the size of NH2-PEG5000-b-CMx-r-OTy polymer nanoparticles were measured for a period of one month at indicated time points (0, 7, 14, 21 and 28 days). During this study, the particles were stored in water at 4°C.
Estimation of Dye leaching out from fluorescent sNPs:
The stability of dye loaded nanoparticles was evaluated by quantifying the absorbance of DiD at 660nm using UV-vis spectrophotometer at day 1 and day 7.
Transmission Electron Microscope (TEM) Study:
The sample used for DLS measurement was dropped onto carbon-coated copper grid. The grid was dried by slow evaporation in air, and then dried separately in vacuum overnight. Images were recorded on a JEOL2000FX electron microscopy operated at 120 kV and at a nominal magnification of 20000X.
Cell culture & stimulation
Immortalized bone-marrow-derived macrophages (iBMDMs) and iBMDMs stably expressing ASC-CFP were obtained as a gift from Dr. Kate Fitzgerald from UMass Medical School Worcester. Immortalized BMDM were generated using J2 transforming retroviruses from C57BL/6 primary BMDMs. They were cultured in DMEM media supplemented with heat-inactivated Fetal Bovine Serum (FBS), penicillin (50 μg/mL) and streptomycin (50 μg/mL). For passaging, 0.25% of trypsin (diluted in PBS) was used.
Before any specific assays, iBMDMs were stimulated with signal 1 and signal 2 required to activate the NLRP3 inflammasome. For signal 1, priming, they were incubated with 50 ng/ml ultrapure LPS for 12–16 hours. These primed iBMDMs were then subjected to signal 2, which involved treatment with nanoparticles or inflammasome-activating drugs. For experimental studies, indicated concentrations of sNPs, and for positive control, Nigericin or SiO2 nanoparticles were used. Signal 2 was treatment was performed for either 24hours or 48hours as indicated in the specific experimental protocols.
IL-1β ELISA assay
iBMDMs seeded at a density of 4 × 104 cells per well in a 96-well plate were further primed with 50 ng/mL of TLR grade LPS overnight. These primed cells were exposed to an appropriate amount of the nanoparticles ranging from 0.05 mg/mL to 1 mg/mL for various times points (12, 24, 48 hours), depending upon the experiment. The cellular supernatants were collected for quantifying IL-1β by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s protocol.
Cytotoxicity assay
Cell viability of the primed and nanoparticle-treated cells was detected by the MTS assay. 8 × 104 total iBMDMs were seeded into 96 well plates, followed overnight 50 ng/mL LPS priming and further nanoparticle (name of Nps) treatment for the indicated time. After treatment, the treated cells were washed with PBS and the media was replaced with phenol red-free basal DMEM followed by incubation in a mixture of MTS and PMS in the ratio 0f 1:20 for 30 minutes. Absorbance was measured using a BioTek plate reader at 490nm O.D., which was further analyzed using Graph Pad software.
Immunoblotting analysis
A density of 8 × 106 iBMDMs were seeded in 10cm2 plates. Cultured cells were primed with 50 ng/mL TLR-grade LPS and further subjected to different nanoparticle (y = 0 to 400) treatments at indicated time points. After treatments, the supernatant was collected and stored after adding 0.1mM of PMSF at −80 degree Celsius under further use. Treated cells were washed with PBS twice and further lysed using protease and phosphatase HALT inhibitor-containing RIPA lysis buffer at 4 degrees Celsius. Total protein extracted in the supernatant obtained after high-speed centrifugation, 14000 rpm, for 15minutes was further quantified using BCA estimation. An equal amount of extracted proteins were separated by SDS-PAGE on a 10–12% polyacrylamide gel and then transferred to a PVDF membrane. Following the blockage with 5% skim milk in TBST, these PVDFs were incubated in primary antibodies of anti-cleaved caspase-1 (1:1000), anti-NLRP3 (1:1000), anti-caspase-1 (1:1000), anti-ASC (1:1000) and anti-b-actin (1:2000) in 1% BSA TBST solution for overnight at 4°C. After washing with TBST, membranes were incubated with HRP-conjugated secondary antibody (1:2000) for 1 hour at room temperature. Primary antibodies specific for caspase-1, active caspase-1, NLRP3 and ASC were purchased from Adipogen. β-actin and secondary antibodies were obtained from Biolegend and Cell Signaling Technology, respectively. Biorad ECL Clarity (Catalog No. 1705061) was used to detect the immunoreactive bands by Biorad ChemiDoc Imaging Systems. To process/analyze the images and quantify the densitometric values, ImageJ software was employed.
Immunofluorescence staining (HCA Assay)
CFP tagged ASC containing iBMDMs (iBMDM ASC-CFP) were seeded at a density of 2 × 105 cells in a glass-bottom 96 well black plate. Adhered cells were then LPS primed overnight. Primed cells were subjected to different nanoparticles treatments, NucRed (2drops in 1 mL media) and Propidium Iodide (conc.) stain; and further imaged at indicated time points using High Content Analysis Microscope at UMass Light Microscopy Core Facility. The images were analyzed and quantified using Nikon NIS-Elements AR (Ver.4.50).
Cellular uptake of nanoparticles
To assess the cellular uptake, CSFE-stained iBMDMs were primed and treated with fluorescent sNPs for indicated time points (3 hours and 9 hours). CSFE staining was according to the user’s protocol. To begin with, 1million CSFE stained cells were seeded in a 12 well plate. Cultured cells were then treated with DiD encapsulated Fluorescent nanoparticles and washed twice with 1X PBS. After washing, they were resuspended in 100 μL of FACS Staining buffer and quantified using ACEA Novocyte flow cytometer. Finally, double-positive cells were gated and analyzed on NovoExpress 1.2.5 software. Percent double-positive cells were plotted using Graph-pad prism.
Lysosomal Rupture Assay: Lysotracker, Acridine Orange
For evaluating lysosomal rupture using lysotracker, 0.2 × 106 iBMDMs were seeded in an 8 well chamber slide (Labtek). After cells adhere to the slide, they were primed with ultrapure LPS for 12 hours, followed by DiD sNPs (y=0 to y=400) treatment for 4 hours. After 4 hours of nanoparticle treatment, cells were washed with PBS twice and stained with 75 nM of LysoTracker™ Red DND 99 for 30 minutes. Nucleus was stained using NucBlue™ Live Ready Probes Reagent™ (Hoechst 33342), 2 drops per mL of media. After all the stainings and washings, warm media was added to the cells, and they were visualized using Nikon CrestV2 spinning disk confocal microscope (CrestV2-2xTIRF) at 60X. Next, the images were adjusted for brightness and contrast and analyzed with NIS Elements 4.6. Lysosomal rupture was quantified by identifying the reduction in percent lysotracker intensity among treatment groups compared to just primed cells. The graph was plotted using Graph Pad Prism software.
For examining lysosomal rupture using acridine orange, iBMDMs were seeded in 12 well plates at a density of 7 × 105 cells/mL. Adhered cells were LPS primed, treated with indicated nanoparticles for 4 hours and then incubated with 2 μg/mL acridine orange (AO) for 2 hours. Treated cells were further washed with PBS twice and finally re-suspended in FACS Staining Buffer (PBS with 3% BSA and 0.1% NaN3). AO-associated orange and green fluorescence intensity were quantified with ACEA Novocyte flow cytometry. Graphs for the reduction in orange fluorescence and increase in green fluorescence were plotted using Graph Pad.
Nanoparticle cellular uptake assessment by flow cytometry and confocal microscopy
iBMDMs were stained with CSFE according to the user’s protocol. For microscopy, 0.2 × 106 CFSE Stained cells were plated in each well of an 8 well chamber slide (Labtek), followed by treatments with fluorescent DiD nanoparticles for the indicated time points. These stained cells were then fixed using 100% methanol and washed twice with 1X PBS. After washing, the cells were counterstained with DAPI, mounted, and imaged using a Nikon CrestV2 spinning disk confocal microscope at 60X and analyzed with NIS Elements 4.6. Images were adjusted for brightness and contrast.
For Flow cytometry, 1million CSFE stained cells were seeded in a 12 well plate. Cultured cells were treated with DiD Fluorescent nanoparticles and washed twice with 1X PBS. After washing, they were resuspended in 100 μL of FACS Staining buffer and quantified using ACEA Novocyte flow cytometer and analyzed on NovoExpress 1.2.5 software.
Intracellular calcium detection using fluo-4 AM
For time-dependent flow cytometry analysis to identify intracellular calcium levels in treated cells, 1 × 106 iBMDMs per well were seeded in a 12 well plate. Adhered cells were then primed with LPS and treated with nanoparticles for indicated time points (4, 12 and 24hours). For positive control the primed cells were incubated with complete media throughout the nanoparticle treatment time points and then treated with nigericin for 20 minutes right before fluo 4 staining. Next, cells were washed twice with PBS and incubated with 1 μM of cell-permeant Fluo-4AM (AM: Acetoxymethyl) in CPBS (PBS with CaCl2 and MgCl2) for 30min at 37°C. y=400 sNPs 12 hours treatment group were also incubated with 10 μM BAPTA-AM cell permeant chelator for 30 minutes in order to evaluate the inhibition in calcium flux. After taking out the cells from the incubator, they were covered in foil and left in the hood for 10 more minutes (at room temperature) before processing for sample acquisition. Next, stained cells were scrapped, washed with PBS twice, resuspended in FACS staining buffer and acquired using ACEA Novocyte flow cytometer. Fluo-4 median fluorescence intensity was analyzed using NovoExpress 1.2.5 software and plotted using Graphpad Prism.
For microscopy studies, iBMDMs were plated at a density of 0.2 × 106 per well in an 8-well labtek chamber slide. They were first primed with ultrapure LPS overnight and then treated with hydrophobic series of sNPs for 12 hours. After signal 1 and signal 2 treatments, these cells were subjected to intracellular calcium staining using 2.5 μM Fluo-4 AM for 30minutes at 37°C. Further, cells were stained with NucBlue for 10 minutes, washed and visualized using Nikon CrestV2 spinning disk confocal microscope after adding warm media.
Determination of mitochondrial ROS generation
To quantify mitochondrial ROS production, 1 × 106 iBMDMs plated in each well (of a 12-well plate) were first primed with LPS for 12 hours, treated with y=0 to y=400 sNPs for indicated time points (4, 12 and 24 hours) and then stained with 1 μM MitoSOX in HBSS at 37°C for 20 minutes. For positive control, the primed cells were treated with nigericin for 30 minutes right before staining and to assess the calcium flux dependent loss of function, the cells were treated with 10 μM BAPTA-AM inhibitor. After MitoSOX treatment, cells were washed with PBS/HBSS, dislodged from the plate using a cell scraper and centrifuged at 2000 rpm for 5minutes to obtain the pellet. The cell pellets of different treatment groups were again washed with PBS to eliminate the residual MitoSOX, resuspended in FACS staining buffer and quantified using ACEA Novocyte flow cytometer.
To visualize mitochondrial ROS production, 1 × 106 iBMDMs per well were LPS primed and treated with nanoparticles for indicated times, then washed twice with PBS and treated with 5 μM of MitoSOX in HBSS at 37°C for 10min. The nuclei were stained using NucBlue, and cells were imaged at 60X oil objective using Nikon CrestV2 spinning disk confocal microscope.
Statistical Analysis
Statistics were computed using ordinary one-way or two-way ANOVA analysis (GraphPad Prism 8). Multiple comparisons were performed using Dunnett’s multiple comparison test. All the results were expressed as mean ± S.E.M (Standard Error of the Mean), and P<0.05 was considered to be significant.
Supplementary Material
ACKNOWLEDGEMENTS:
General:
We would like to acknowledge the support and assistance of the Biophysical Characterization Core Facility, Light Microscopy Core Facility, and Animal Imaging Facility at the Institute for Applied Life Sciences (IALS), and the Electron Microscopy Core Facility at the University of Massachusetts Amherst. A special thanks to Dr. James Chambers for helping in optimizing microscopy assays and their analysis. We would like to thank Prof. Kate Fitzgerald, UMass Medical School, Worcester for kindly donating the iBMDM and ASC-CFP iBMDMs cell line. We would also like to thank Jacob Ullom, Noorul Shaheen and Hayat Ranjini for their help in generating the data. The general figures were created with Biorender.com.
Funding:
This work was financially supported by the American Cancer Society Research Scholar Grant (RSG-19-009-01-CDD), Melanoma Research Alliance Young Investigator Award (510283) and Cancer Research Institute Technology Impact Award (118-1501) to A. K. and NIGMS of the NIH grant (GM-136395) to ST.
ABBREVIATIONS
- NAMPs
Nanoparticle-Associated-Molecular-Patterns
- PAMPs
Pathogen-associated-molecular-patterns
- DAMPs
Damage-associated-molecular-patterns
- NLR
NOD-like Receptor
- NLRP3
nucleotide-binding domain (NOD)-like receptor protein 3
- TLR
Toll-like Receptor
- PRRs
Pattern Recognition Receptor
- ASC
Apoptosis-associated speck like protein
- sNPs
Supramolecular polymeric nanoparticles
- IL-1β
Interleukin-1β
- HCA
High Content Analysis
- iBMDMs
Immortalized bone-marrow-derived-macrophages
- ROS
Reactive Oxygen Species
- Fluo-4AM
AM: Acetoxymethyl
- LPS
Lipopolysaccharide
- CFSE
Carboxyfluorescein succinimidyl ester
- MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
- TNF-α
Tumor Necrosis Factor
- IFN-γ
Interferon-gamma
Footnotes
Supporting Information:
The supporting information is available free of charge at ACS.
Competing Interests: The authors declare no competing interests.
REFERENCES
- 1.Martinon F, Burns K, Tschopp J, The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of proIL-β. Molecular Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Bergsbaken T, Fink SL, Cookson BT, Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 7, 99–109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Wang H, Kouadir M, Song H, Shi F, Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death & Disease 10, 128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley N, Jeltema D, Duan Y, He Y, The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. International Journal of Molecular Sciences 20, 1–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E, Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov 17, 588–606 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Yuan He HH, Gabriel Núñez, Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends in Biochemical Sciences 41, 1012–1021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries F, Fitzgerald KA, Assembling the Inflammasome, Piece by Piece. The Journal of Immunology 203, 1093–1094 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Rathinam VAK, Fitzgerald KA, Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 165, 792–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rathinam VAK, Vanaja SK, Fitzgerald KA, Regulation of inflammasome signaling. Nature Immunology 13, 333–342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyano DF, Liu Y, Peer D, Rotello VM, Modulation of Immune Response Using Engineered Nanoparticle Surfaces. Small 12, 76–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boraschi D, Italiani P, Palomba R, Decuzzi P, Duschl A, Fadeel B, Moghimi SM, Nanoparticles and innate immunity: new perspectives on host defence. Semin Immunol 34, 33–51 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Muhammad Q, Jang Y, Kang SH, Moon J, Kim WJ, Park H, Modulation of immune responses with nanoparticles and reduction of their immunotoxicity. Biomater Sci 8, 1490–1501 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Grimaldi AM, Incoronato M, Salvatore M, Soricelli A, Nanoparticle-based strategies for cancer immunotherapy and immunodiagnostics. Nanomedicine 12, 2349–2365 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Syed S, Zubair A, Frieri M, Immune response to nanomaterials: implications for medicine and literature review. Curr Allergy Asthma Rep 13, 50–57 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kononenko V, Narat M, Drobne D, Nanoparticle interaction with the immune system. Arh Hig Rada Toksikol 66, 97–108 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Shirasuna K, Karasawa T, Takahashi M, Exogenous nanoparticles and endogenous crystalline molecules as danger signals for the NLRP3 inflammasomes. Journal of Cellular Physiology 234, 5436–5450 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Luan J, Chen W, Fan J, Nan Y, Wang Y, Liang Y, Meng G, Ju D, Mesoporous silica nanoparticles induced hepatotoxicity via NLRP3 inflammasome activation and caspase-1-dependent pyroptosis. Nanoscale 10, 9141–9152 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Baron L, Gombault A, Fanny M, Villeret B, Savigny F, Guillou N, Panek C, Le Bert M, Lagente V, Rassendren F, Riteau N, Couillin I, The NLRP3 inflammasome is activated by nanoparticles through ATP, ADP and adenosine. Cell Death and Disease 6, 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E, Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9, 847–856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez DM, Urcuqui-Inchima S, Hernandez JC, Silica nanoparticles induce NLRP3 inflammasome activation in human primary immune cells. Innate Immunity 23, 697–708 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Sasabe E, Tomomura A, Kitamura N, Yamamoto T, Metal nanoparticles-induced activation of NLRP3 inflammasome in human oral keratinocytes is a possible mechanism of oral lichenoid lesions. Toxicology in Vitro 62, 104663 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Harizaj A, Hauwermeiren FV, Stremersch S, Rycke RD, Keersmaecker HD, Brans T, Fraire JC, Grauwen K, Smedt SCD, Lentacker I, Lamkanfi M, Braeckmans K, Nanoparticle-sensitized photoporation enables inflammasome activation studies in targeted single cells. Nanoscale 13, 6592–6604 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Hu Q, Zhao F, Guo F, Wang C, Fu Z, Polymeric Nanoparticles Induce NLRP3 Inflammasome Activation and Promote Breast Cancer Metastasis. Macromolecular Bioscience 17, 1700273 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Reisetter AC, Stebounova LV, Baltrusaitis J, Powers L, Gupta A, Grassian VH, Monick MM, Induction of Inflammasome-dependent Pyroptosis by Carbon Black Nanoparticles *. Journal of Biological Chemistry 286, 21844–21852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang E-J, Kim S, Kim JS, Choi I-H, Inflammasome formation and IL-1β release by human blood monocytes in response to silver nanoparticles. Biomaterials 33, 6858–6867 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Peeters PM, Perkins TN, Wouters EF, Mossman BT, Reynaert NL, Silica induces NLRP3 inflammasome activation in human lung epithelial cells. Particle and Fibre Toxicology 10, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Z, Zhai Y, Liang S, Mori Y, Han R, Sutterwala FS, Qiao L, TRPM2 links oxidative stress to NLRP3 inflammasome activation. Nature communications 4, 1611–1611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y-H, Chang LW, Lin P, Metal-Based Nanoparticles and the Immune System: Activation, Inflammation, and Potential Applications. BioMed Research International 2015, 143720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun B, Wang X, Ji Z, Li R, Xia T, NLRP3 inflammasome activation induced by engineered nanomaterials. Small 9, 1595–1607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma B, McLeland CB, Potter TM, Stern ST, Adiseshaiah PP, Assessing NLRP3 Inflammasome Activation by Nanoparticles. Methods Mol Biol 1682, 135–147 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Yang M, Flavin K, Kopf I, Radics G, Hearnden CHA, McManus GJ, Moran B, Villalta-Cerdas A, Echegoyen LA, Giordani S, Lavelle EC, Functionalization of Carbon Nanoparticles Modulates Inflammatory Cell Recruitment and NLRP3 Inflammasome Activation. Small 9, 4194–4206 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Mishra AR, Zheng J, Tang X, Goering PL, Silver nanoparticle-induced autophagic-Lysosomal disruption and NLRP3-inflammasome activation in HepG2 cells is size-dependent. Toxicological Sciences 150, 473–487 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Sha R, Yang L, Zhao X, Zhu Y, Gao J, Zhang Y, Wen LP, Impact of Morphology on Iron Oxide Nanoparticles-Induced Inflammasome Activation in Macrophages. ACS Applied Materials and Interfaces 10, 41197–41206 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Silva AL, Peres C, Conniot J, Matos AI, Moura L, Carreira B, Sainz V, Scomparin A, Satchi-Fainaro R, Preat V, Florindo HF, Nanoparticle impact on innate immune cell pattern-recognition receptors and inflammasomes activation. Semin Immunol 34, 3–24 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Palomaki J, Valimaki E, Sund J, Vippola M, Clausen PA, Jensen KA, Savolainen K, Matikainen S, Alenius H, Long, needle-like carbon nanotubes and asbestos activate the NLRP3 inflammasome through a similar mechanism. ACS nano 5, 6861–6870 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lunov O, Syrovets T, Loos C, Nienhaus GU, Mailänder V, Landfester K, Rouis M, Simmet T, Amino-functionalized polystyrene nanoparticles activate the NLRP3 inflammasome in human macrophages. ACS Nano 5, 9648–9657 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Vaine CA, Patel MK, Zhu J, Lee E, Finberg RW, Hayward RC, Kurt-Jones EA, Tuning Innate Immune Activation by Surface Texturing of Polymer Microparticles: The Role of Shape in Inflammasome Activation. The Journal of Immunology 190, 3525–3532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baljon JJ, Dandy A, Wang-Bishop L, Wehbe M, Jacobson ME, Wilson JT, The efficiency of cytosolic drug delivery using pH-responsive endosomolytic polymers does not correlate with activation of the NLRP3 inflammasome. Biomaterials Science 7, 1888–1897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabolli V, Lison D, Huaux F, The complex cascade of cellular events governing inflammasome activation and IL-1β processing in response to inhaled particles. Particle and Fibre Toxicology 13, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manna S, Howitz WJ, Oldenhuis NJ, Eldredge AC, Shen J, Nihesh FN, Lodoen MB, Guan Z, Esser-Kahn AP, Immunomodulation of the NLRP3 Inflammasome through Structure-Based Activator Design and Functional Regulation via Lysosomal Rupture. ACS Central Science 4, 985–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang YJ, Hung KC, Hung HS, Hsu SH, Modulation of Macrophage Phenotype by Biodegradable Polyurethane Nanoparticles: Possible Relation between Macrophage Polarization and Immune Response of Nanoparticles. ACS Appl Mater Interfaces 10, 19436–19448 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Chang CH, Jiang J, Liu Q, Liao YP, Lu J, Li L, Liu X, Kim J, Ahmed A, Nel AE, Xia T, The Crystallinity and Aspect Ratio of Cellulose Nanomaterials Determine Their Pro-Inflammatory and Immune Adjuvant Effects In Vitro and In Vivo. Small 15, e1901642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Despres HW, Sabra A, Anderson P, Hemraz UD, Boluk Y, Sunasee R, Ckless K, Mechanisms of the immune response cause by cationic and anionic surface functionalized cellulose nanocrystals using cell-based assays. Toxicol In Vitro 55, 124–133 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Shima F, Akagi T, Akashi M, The hydrophobic effect of nanoparticles composed of amphiphilic poly(γ-glutamic acid) on the degradability of the encapsulated proteins. Biomaterials Science 2, 1419–1425 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Moyano DF, Goldsmith M, Solfiell DJ, Landesman-Milo D, Miranda OR, Peer D, Rotello VM, Nanoparticle Hydrophobicity Dictates Immune Response. Journal of the American Chemical Society 134, 3965–3967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Hardie J, Zhang X, Rotello VM, Effects of engineered nanoparticles on the innate immune system. Seminars in Immunology 34, 25–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B, Xie J, Yuan Z, Jain P, Lin X, Wu K, Jiang S, Mitigation of Inflammatory Immune Responses with Hydrophilic Nanoparticles. Angewandte Chemie International Edition 57, 4527–4531 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Li C, Zhou J, Wu Y, Dong Y, Du L, Yang T, Wang Y, Guo S, Zhang M, Hussain A, Xiao H, Weng Y, Huang Y, Wang X, Liang Z, Cao H, Zhao Y, Liang X-J, Dong A, Huang Y, Core Role of Hydrophobic Core of Polymeric Nanomicelle in Endosomal Escape of siRNA. Nano Letters 21, 3680–3689 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Wang S, Guo H, Li Y, Li X, Penetration of nanoparticles across a lipid bilayer: effects of particle stiffness and surface hydrophobicity. Nanoscale 11, 4025–4034 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Sun J, Zhang L, Wang J, Feng Q, Liu D, Yin Q, Xu D, Wei Y, Ding B, Shi X, Jiang X, Tunable rigidity of (polymeric core)-(lipid shell) nanoparticles for regulated cellular uptake. Adv Mater 27, 1402–1407 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Demento SL, Eisenbarth SC, Foellmer HG, Platt C, Caplan MJ, Mark Saltzman W, Mellman I, Ledizet M, Fikrig E, Flavell RA, Fahmy TM, Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine 27, 3013–3021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp FA, Ruane D, Claass B, Creagh E, Harris J, Malyala P, Singh M, O’Hagan DT, Pétrilli V, Tschopp J, O’Neill LAJ, Lavelle EC, Uptake of particulate vaccine adjuvants by dendritic cells activates the NALP3 inflammasome. Proceedings of the National Academy of Sciences of the United States of America 106, 870–875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice-Ficht AC, Arenas-Gamboa AM, Kahl-McDonagh MM, Ficht TA, Polymeric particles in vaccine delivery. Curr Opin Microbiol 13, 106–112 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Gao J, Wu P, Fernandez A, Zhuang J, Thayumanavan S, Cellular AND Gates: Synergistic Recognition to Boost Selective Uptake of Polymeric Nanoassemblies. Angewandte Chemie International Edition 59, 10456–10460 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Gao J, Wu P, Fernandez A, Zhuang J, Thayumanavan S, Cellular AND Gates: Synergistic Recognition to Boost Selective Uptake of Polymeric Nanoassemblies. Angew Chem Int Ed Engl 59, 10456–10460 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Cao R, Gao J, Thayumanavan S, Dinsmore AD, Triggered interactions between nanoparticles and lipid membranes: design principles for gel formation or disruption-and-release. Soft Matter 17, 7069–7075 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Gao J, Dutta K, Zhuang J, Thayumanavan S, Cellular- and Subcellular-Targeted Delivery Using a Simple All-in-One Polymeric Nanoassembly. Angew Chem Int Ed Engl 59, 23466–23470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez A, Zentner CA, Shivrayan M, Samson E, Savagatrup S, Zhuang J, Swager TM, Thayumanavan S, Programmable Emulsions via Nucleophile-Induced Covalent Surfactant Modifications. Chemistry of Materials 32, 4663–4671 (2020). [Google Scholar]
- 59.De Nardo D, Kalvakolanu DV, Latz E, in Macrophages: Methods and Protocols, Rousselet G, Ed. (Springer New York, New York, NY, 2018), vol. 1784, pp. 35–49. [DOI] [PubMed] [Google Scholar]
- 60.Zito G, Buscetta M, Cimino M, Dino P, Bucchieri F, Cipollina C, Cellular Models and Assays to Study NLRP3 Inflammasome Biology. International journal of molecular sciences 21, 4294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko JW, Shin NR, Je-Oh L, Jung TY, Moon C, Kim TW, Choi J, Shin IS, Heo JD, Kim JC, Silica dioxide nanoparticles aggravate airway inflammation in an asthmatic mouse model via NLRP3 inflammasome activation. Regul Toxicol Pharmacol 112, 104618 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Yazdi AS, Guarda G, Riteau N, Drexler SK, Tardivel A, Couillin I, Tschopp J, Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1alpha and IL-1beta. Proceedings of the National Academy of Sciences of the United States of America 107, 19449–19454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marzaioli V, Groß CJ, Weichenmeier I, Schmidt-weber CB, Gutermuth J, Groß O, Alessandrini F, Specific Surface Modifications of Silica Nanoparticles Diminish Inflammasome Activation and In Vivo Expression of Selected Inflammatory Genes. nanomaterials 7, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marzaioli V, Gross CJ, Weichenmeier I, Schmidt-Weber CB, Gutermuth J, Gross O, Alessandrini F, Specific Surface Modifications of Silica Nanoparticles Diminish Inflammasome Activation and In Vivo Expression of Selected Inflammatory Genes. Nanomaterials (Basel) 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim ST, Saha K, Kim C, Rotello VM, The role of surface functionality in determining nanoparticle cytotoxicity. Accounts of chemical research 46, 681–691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franklin BS, Bossaller L, Nardo DD, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, Mangan MS, Zimmer S, Monks BG, Fricke M, Schmidt RE, Espevik T, Jones B, Jarnicki AG, Hansbro PM, Busto P, Marshak-Rothstein A, Hornemann S, Aguzzi A, Kastenmüller W, Latz E, The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nature Immunology 15, 727–737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES, The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ 14, 1590–1604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt FI, Lu A, Chen JW, Ruan J, Tang C, Wu H, Ploegh HL, A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J Exp Med 213, 771–790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu T, Yamaguchi Y, Shirasaki Y, Shikada K, Yamagishi M, Hoshino K, Kaisho T, Takemoto K, Suzuki T, Kuranaga E, Ohara O, Miura M, Single-cell imaging of caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Rep 8, 974–982 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Sager TM, Wolfarth M, Leonard SS, Morris AM, Porter DW, Castranova V, Holian A, Role of engineered metal oxide nanoparticle agglomeration in reactive oxygen species generation and cathepsin B release in NLRP3 inflammasome activation and pulmonary toxicity. Inhal Toxicol 28, 686–697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jessop F, Hamilton RF, Rhoderick JF, Fletcher P, Holian A, Phagolysosome acidification is required for silica and engineered nanoparticle-induced lysosome membrane permeabilization and resultant NLRP3 inflammasome activity. Toxicology and Applied Pharmacology 318, 58–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yaron JR, Gangaraju S, Rao MY, Kong X, Zhang L, Su F, Tian Y, Glenn HL, Meldrum DR, K(+) regulates Ca(2+) to drive inflammasome signaling: dynamic visualization of ion flux in live cells. Cell death & disease 6, e1954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J, Wang X, Mei K-C, Chang CH, Jiang J, Liu X, Liu Q, Guiney LM, Hersam MC, Liao Y-P, Meng H, Xia T, Lateral size of graphene oxide determines differential cellular uptake and cell death pathways in Kupffer cells, LSECs, and hepatocytes. Nano Today 37, 101061 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holley CL, Schroder K, The rOX-stars of inflammation: links between the inflammasome and mitochondrial meltdown. Clinical and Translational Immunology 9, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coll RC, Holley CL, Schroder K, Mitochondrial DNA synthesis fuels NLRP3 activation. Cell Res 28, 1046–1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zha Z, Li J, Ge Z, Endosomal-Escape Polymers Based on Multicomponent Reaction-Synthesized Monomers Integrating Alkyl and Imidazolyl Moieties for Efficient Gene Delivery. ACS Macro Letters 4, 1123–1127 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Manganiello MJ, Cheng C, Convertine AJ, Bryers JD, Stayton PS, Diblock copolymers with tunable pH transitions for gene delivery. Biomaterials 33, 2301–2309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murthy N, Robichaud JR, Tirrell DA, Stayton PS, Hoffman AS, The design and synthesis of polymers for eukaryotic membrane disruption. Journal of controlled release : official journal of the Controlled Release Society 61, 137–143 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Carmona-Ribeiro AM, Perez-Betancourt Y, Cationic Nanostructures for Vaccines Design. Biomimetics (Basel) 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


