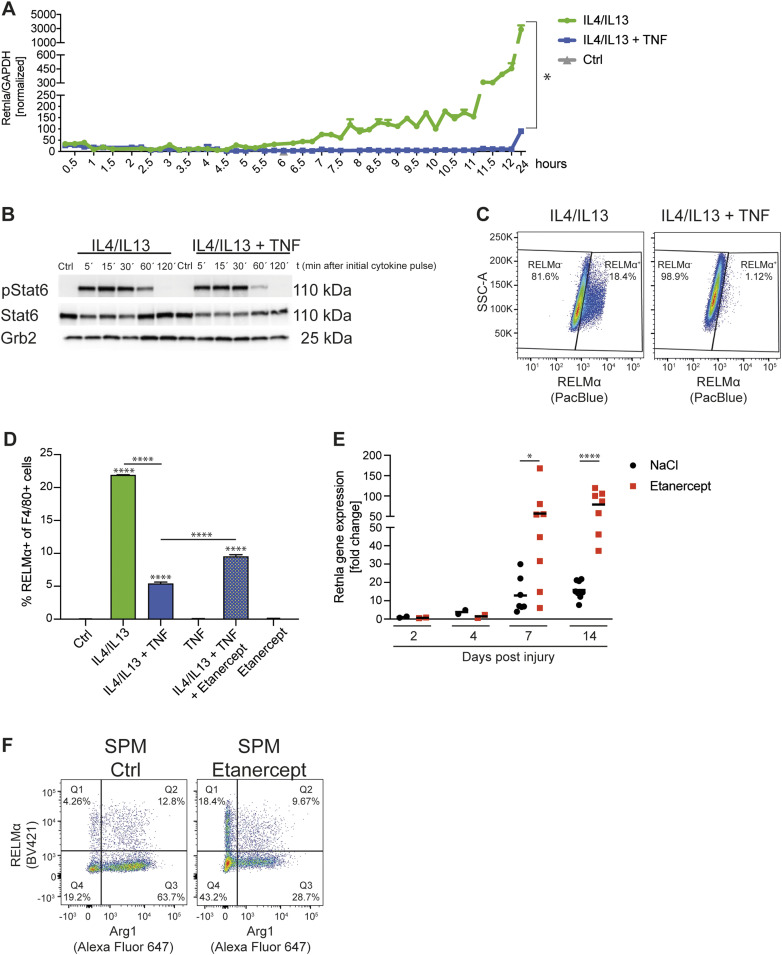
Figure 1. TNF inhibits M2 gene expression.
(A) RNA from wild-type BMDMs left untreated (Ctrl), stimulated with IL4/IL13 or IL4/IL13 + TNF was used for qRT-PCR. Data shown are the mean fold-increase compared with the Ctrl group. (B) Wild-type BMDMs, which were treated with solvent (Ctrl), IL4/IL13 or IL4/IL13 + TNF for 5, 15, 30, 60, or 120 min, were used to isolate whole-cell lysates and analyzed by Western blotting for the indicated proteins. (C) Representative plots of wild-type BMDMs stimulated with IL4/IL13 or IL4/IL13 + TNF and percentages of RELMα+ of F4/80+ macrophages were determined by flow cytometry analysis. (D) Wild-type BMDMs were stimulated with IL4/IL13, IL4/IL13 + TNF and/or Etanercept and percentages of RELMα+ macrophages were determined by flow cytometry analysis. Data represent three independent experiments. (E) CD45+CD11b+F4/80+ macrophages from skin excisional wounds were sorted, RNA was extracted and analyzed by qRT-PCR. Data are representative for two independent experiments. (F) Small peritoneal macrophages were analyzed after in vivo injection of solvent or Etanercept. Percentages of Arg1+ and RELMα+ cells were analyzed by flow cytometry analysis. See also Fig S1. All values are means ± SEM; *P < 0.05; ****P < 0.0001. Statistically significant differences were determined by one-way ANOVA with Tukey’s correction or by paired t test. If not indicated otherwise superscripts show statistical significance compared with the control group. n = 3 biological replicates.