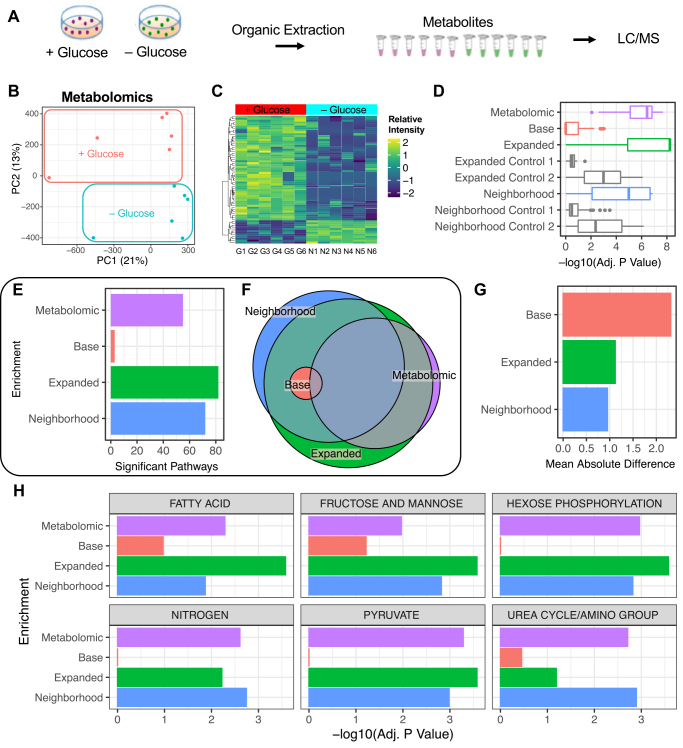
Fig. 3.
Experimental validation of metabolomic changes in breast cancer cell line.A, experimental workflow of metabolomic profiling of glucose-starved cell line. B, PCA dimensionality reduction of unbiased metabolomic profiles showing partitioning of Glucose versus No Glucose samples. C, heatmap of top differential annotated metabolites identified using a decoy adduct strategy. D, global pathway enrichment results comparing the global significance of phospho/proteomic pathways with metabolomics-based analysis using the same Base, Expanded, Neighborhood, and Control gene sets. E, number of significant metabolic pathways (Adj. p Value < 0.05) enriched in the metabolomic and phospho/proteomic data using the Base, Expanded, and Neighborhood gene sets. F, overlap of significant (Adj. p Value < 0.05) metabolic pathways enriched in the metabolomic data and phospho/proteomic data using the Base, Expanded, and Neighborhood gene sets. G, mean absolute difference of pathway significance between the Metabolomic and phospho/proteomic enrichment results. H, significance of select metabolic pathways across the metabolomic and phospho/proteomic profiles. PCA, principal component analysis.