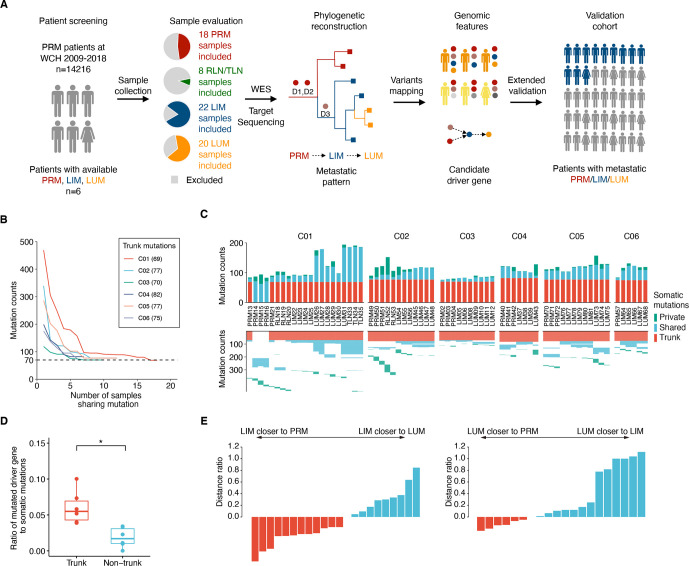
Figure 1.
Tracking tumour evolution of distant metastases in colorectal cancers. (A) Study design schematic. samples from primary tumour (PRM), regional lymph node metastasis (RLN), liver metastasis (LIM), lung metastasis (LUM), thoracic lymph node metastasis (TLN) and normal tissue from six patients with colorectal cancer were selected based on pathological evaluation and sent for whole-exome sequencing (WES). Phylogenetic trees were reconstructed to infer the metastatic seeding pattern of each case. D1–D3 stand for different driver mutations acquired during metastatic evolution. Multiregion genomic data from each patient were jointly analysed. The candidate novel features were validated in two independent cohorts. (B) Mutation counts shared by at least n tumour samples in each patient. (C) The total number of somatic mutations is visualised as stacked bars classified by private mutations, shared mutations (recur in at least two tumour samples) and trunk mutations (shared by all tumour samples) in each individual (top). The trunk and shared mutations were clustered to illuminate the phylogenetic relationship of each mutation among tumour samples (bottom). (D) Fractions of driver gene mutations are higher in trunk mutations compared with non-trunk mutations. (*p<0.05, two-sided paired t-test) (E) (left) Most LIM are more closely related to the PRM than to LUM. The plot shows the d(LIM to PRM)/d(LIM to LUM) – 1. (The distance of each LIM to its closest PRM, divided by its distance to its closest LUM, minus one) (right) Analogous plot for LUM, showing d (LUM to PRM)/d(LUM to LIM) – 1.