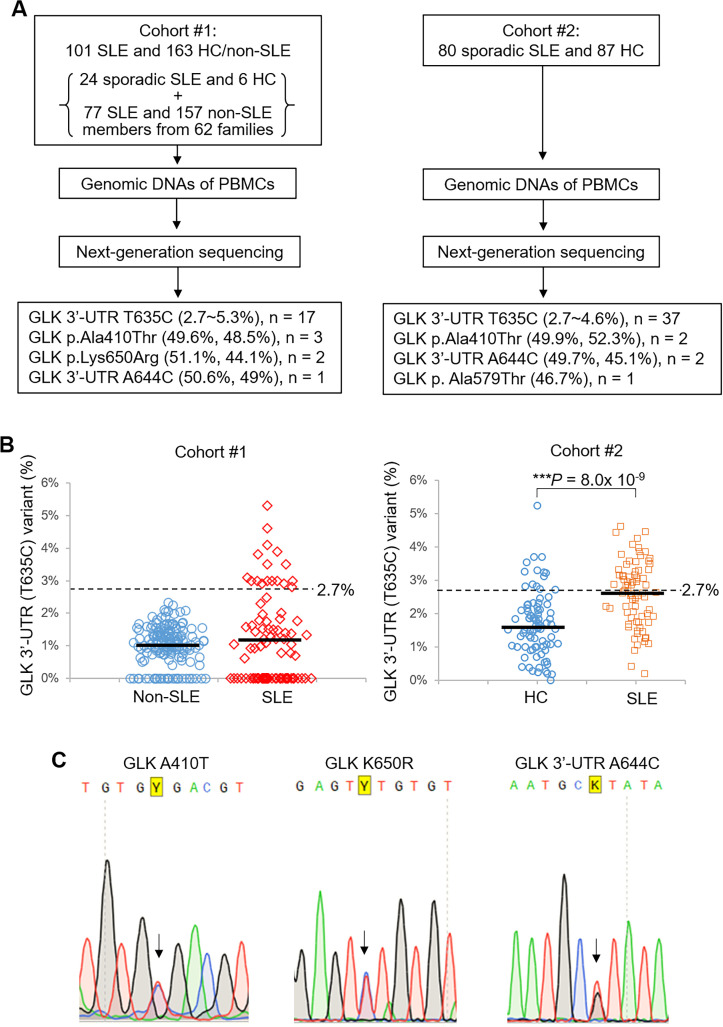
Figure 1.
GLK somatic and germline variants occur in PBMCs of SLE patients. (A) Schematic diagram of the screening design to identify GLK gene variants in patients with SLE by next-generation sequencing. (B) The variant frequency of GLK 3′-UTR (T635C) variant in 163 individuals without SLE (6 HCs and 157 members without SLE from individual families) and 101 patients with SLE (sporadic and familial) from Cohort #1 (left panel). The value 2.7% was mean plus 3SD (1.0% + 3 × 0.58%=2.74%) of 3′-UTR (T635C) variant frequencies in the group without SLE. The frequency of GLK 3′-UTR (T635C) variant in 87 HCs and 80 patients with SLE (all sporadic) from Cohort #2 (right panel). Bars denote means of variant frequency. ***P<0.0001 (two-tailed Student’s t-test). (C) Sanger-sequencing chromatograms for heterozygous variants at GLK (A410T), (K650R) and 3′-UTR (A644C). Arrows indicate the bases with distinct nucleotides. Y denotes mixed bases of C and T nucleotides; K denotes mixed bases of G and T nucleotides. HCs, healthy controls; PBMCs, peripheral blood mononuclear cells; SLE, systemic lupus erythematosus; UTR, untranslated region.