Abstract
In the present study, orexinergic cell bodies within the brains of rhythmic and arrhythmic circadian chronotypes from three species of African mole rat (Highveld mole rat—Cryptomys hottentotus pretoriae, Ansell’s mole rat—Fukomys anselli and the Damaraland mole rat—Fukomys damarensis) were identified using immunohistochemistry for orexin-A. Immunopositive orexinergic (Orx+) cell bodies were stereologically assessed and absolute numbers of orexinergic cell bodies were determined for the distinct circadian chronotypes of each species of mole rat examined. The aim of the study was to investigate whether the absolute numbers of identified orexinergic neurons differs between distinct circadian chronotypes with the hypothesis of elevated hypothalamic orexinergic neurons in the arrhythmic chronotypes compared with the rhythmic chronotypes. We found statistically significant differences between the circadian chronotypes of F. anselli, where the arrhythmic group had higher mean numbers of hypothalamic orexin neurons compared with the rhythmic group. These differences were observed when the raw data was compared and when the raw data was corrected for body mass (Mb) and brain mass (Mbr). For the two other species investigated, no significant differences were noted between the chronotypes, although a statistically significant difference was noted between all rhythmic and arrhythmic individuals of the current study when the counts of orexin neurons were corrected for Mb—the arrhythmic individuals had larger numbers of orexin cells.
Keywords: orexin, hypocretin, circadian rhythmicity, sleep
It is well known that mole rats, subterranean, visually regressed rodents, have unusual patterns of circadian rhythmicity (Lovegrove and Papenfus, 1995; Lovegrove and Muir, 1996; Oosthuizen et al., 2003). Locomotor activity studies have shown that within a species of mole rat, there are individuals that have a rhythmic chronotype, and others that are distinctly arrhythmic, and this is seen in both social and solitary species of mole rat (Oosthuizen et al., 2003). A study of sleep patterns in the giant Zambian mole rat (Cryptomys mechowi) showed some differences in sleep patterns between rhythmic and arrhythmic chronotypes of this species, where arrhythmic individuals spend more time in waking with a longer average duration of a waking episode, and less time in non-rapid eye movement sleep (NREM) sleep with a shorter average duration of a NREM episode, although with a greater NREM sleep intensity (Bhagwandin et al., 2011b).
The neurotransmitter orexin is known to promote wakefulness, particularly during motor activities (Estabrooke et al., 2001; Mileykovskiy et al., 2005; Lee et al., 2005; Saper et al., 2005). Orexinergic neurons, which are restricted to the hypothalamus, have ascending projections to the cerebral cortex and descending projections to the cholinergic (basal forebrain, lateral dorsal tegmental nucleus, pedunculopontine nucleus) and monoaminergic (locus coeruleus, ventral tegmental area, raphe) nuclear groups involved in the sleep–wake cycle (Peyron et al., 1998; Chemelli et al., 1999; Baumann and Bassetti, 2005). Loss of orexinergic neurons has been associated with narcolepsy (Chemelli et al., 1999; Lin et al., 1999; Siegel, 1999), narcolepsy with cataplexy in humans (Peyron et al., 2000; Thannickal et al., 2000), narcolepsy without cataplexy in humans (Thannickal et al., 2009), and the narcoleptic like symptoms in patients with Parkinson’s disease (Thannickal et al., 2007).
It is also known that orexins are involved in appetite regulation through activation of the leptin sensitive neurons of the arcuate nucleus and its subsequent feeding associated signaling to orexinergic neurons of the lateral hypothalamus (LH) and henceforth interaction between LH and the hypothalamic ventromedial nucleus (VMH) (Stellar, 1954; Rodgers et al., 2002; Sakurai, 2003, 2005). Several studies have demonstrated increased orexinergic activity during states of vigilance and foraging and as a result have paired waking with hunger sensations and optimal feeding (Sakurai et al., 1998; Edwards et al., 1999; Haynes et al., 1999; VanItallie, 2006). Conversely, anorexinergic activity has been associated with sleep states, thereby pairing satiety with sleep (Rodgers et al., 2002; Nicolaidis, 2006; VanItallie, 2006). These studies are indicative of a coordinated orexinergic control of appetite regulation with the sleep–wake cycle that is responsive to the suprachiasmatic nucleus (SCN) circadian clock and nutritional status (Sakurai, 2005; VanItallie, 2006), plus, it has been suggested that disruption of the circadian cycle and sleep deprivation can affect energy balance resulting in changes in body composition (Taheri et al., 2004; VanItallie, 2006).
Given this scenario on the action of the orexinergic neurons, and the clearly distinct circadian chronotypes of the African mole rats, along with differences in the neuropeptide population of the SCN of the mole rats (Negroni et al., 2003), it was thought that an investigation of orexinergic neuronal numbers may yield results of interest in relation to function, especially in the mole rats. In the current study, orexinergic neuronal numbers of three species of African mole rat, the Highveld mole rat (Cryptomys hottenttotus pretoriae [CHP]), Ansell’s mole rat (Fukomys anselli [FA]) and the Damaraland mole rat (Fukomys damarensis [FD]) were quantified. It should be noted that Ansell’s and Damaraland mole rats were previously considered to be members of the genus Cryptomys. All three species of mole rat have a reduced visual system, live a subterranean lifestyle, and have unusual patterns of circadian rhythmicity (Lovegrove and Papenfus, 1995; Lovegrove and Muir, 1996; Oelschläger et al., 2000; Cernuda-Cernuda et al., 2003; Negroni et al., 2003; Oosthuizen et al., 2003; Gutjahr et al., 2004; Nemec et al., 2004, 2008). Brains from circadian distinct rhythmic and arrhythmic individuals of each species were examined. The unusual circadian patterns of individuals within a species provide interesting models to examine whether the absolute numbers of identified orexinergic neurons differs between circadian chronotypes among the species examined.
MATERIALS AND METHODS
A total of 18 brains (15 males and 3 females—see Table 1) from three species of wild caught African mole rat, the Highveld mole rat CHP (average body mass: 127.9 g; average brain mass: 1.5 g), Ansell’s mole rat FA (average body mass: 84.5 g; average brain mass: 1.2 g), and the Damaraland mole rat FD (average body mass: 107.8 g; average brain mass: 1.8 g) were used in this study. For the purposes of the current study, three rhythmic and three arrhythmic individuals per species were used. A single mole rat was housed in an enclosure (30×25 cm2) and wood shavings were used to line the floor. The mole rats used in the present study were caught from wild populations in South Africa under permission and supervision from the appropriate wildlife directorates. All animals were treated and used according to the guidelines of the University of Pretoria Animal Ethics Committee, which parallel those of the NIH for the care and use of animals in scientific experimentation.
Table 1.
Stereological parameters used for rhythmic and arrhythmic individuals of Cryptomys hottentotus pretoriae, Fukomys anselli, and F. damarensis
| Animal ID |
Circadian chronotype |
Counting frame size (μm) |
Sampling grid size (μm) |
Cut thickness (μm) |
Average species mounted thickness (μm) |
Vertical guard zones (top and bottom, μm) |
Section interval |
Number of sampling sites |
Average CE (Gunders, m=0) |
Average CE (Gunders, m=1) |
|---|---|---|---|---|---|---|---|---|---|---|
| C. hottentotus pretoriae | ||||||||||
| CHP2 | R | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 263 | 0.13 | 0.06 |
| CHP8 | R | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 307 | 0.04 | 0.04 |
| CHP12 | R | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 344 | 0.14 | 0.05 |
| CHP1 | AR | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 421 | 0.04 | 0.04 |
| CHP3 | AR | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 347 | 0.06 | 0.04 |
| CHP7 | AR | 275×185×12 | 400×300 | 50 | 20 | 5 | 2 | 358 | 0.05 | 0.04 |
| F. anselli | ||||||||||
| FA2 | R | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 521 | 0.07 | 0.05 |
| FA3 | R | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 438 | 0.09 | 0.05 |
| FA7 | R | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 352 | 0.10 | 0.05 |
| FA4 | AR | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 446 | 0.12 | 0.05 |
| FA5 | AR | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 458 | 0.07 | 0.04 |
| FA6 | AR | 275×185×10 | 400×300 | 50 | 14 | 5 | 2 | 348 | 0.04 | 0.04 |
| F. damarensis | ||||||||||
| FD4 | R | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 331 | 0.05 | 0.04 |
| FD6 | R | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 476 | 0.05 | 0.04 |
| FD7 | R | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 376 | 0.08 | 0.04 |
| FD1 | AR | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 436 | 0.04 | 0.04 |
| FD8 | AR | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 443 | 0.04 | 0.04 |
| FD13 | AR | 275×185×10 | 400×300 | 50 | 15 | 5 | 2 | 371 | 0.08 | 0.05 |
Determination of rhythmicity patterns
Infrared detectors were used to detect activity patterns in the mole rats. The mole rats were exposed to differing light conditions that included, light (L) and dark (D) cycles, DD cycles and LL cycles. Initially, the experimental protocol started with a 12 L: 12 D cycle, and once entrainment was achieved, the light cycle was switched to constant darkness for 1 month. Recorded behavioral activity data were analyzed using Actiview and Clocklab software which were also used to generate actograms. The sensitivity of the y-axis remained the same during analysis of each mole rat. For the purposes of the current study, it was of importance to determine whether the mole rats were able to re-entrain to light cycles and switch their activity according to the light. If a mole rat was able to re-entrain, it was deemed “rhythmic” and the converse were applied to “arrhythmic” mole rats (Fig. 1). For details regarding the specifics of circadian rhythm determination see Oosthuizen et al. (2003).
Fig. 1.
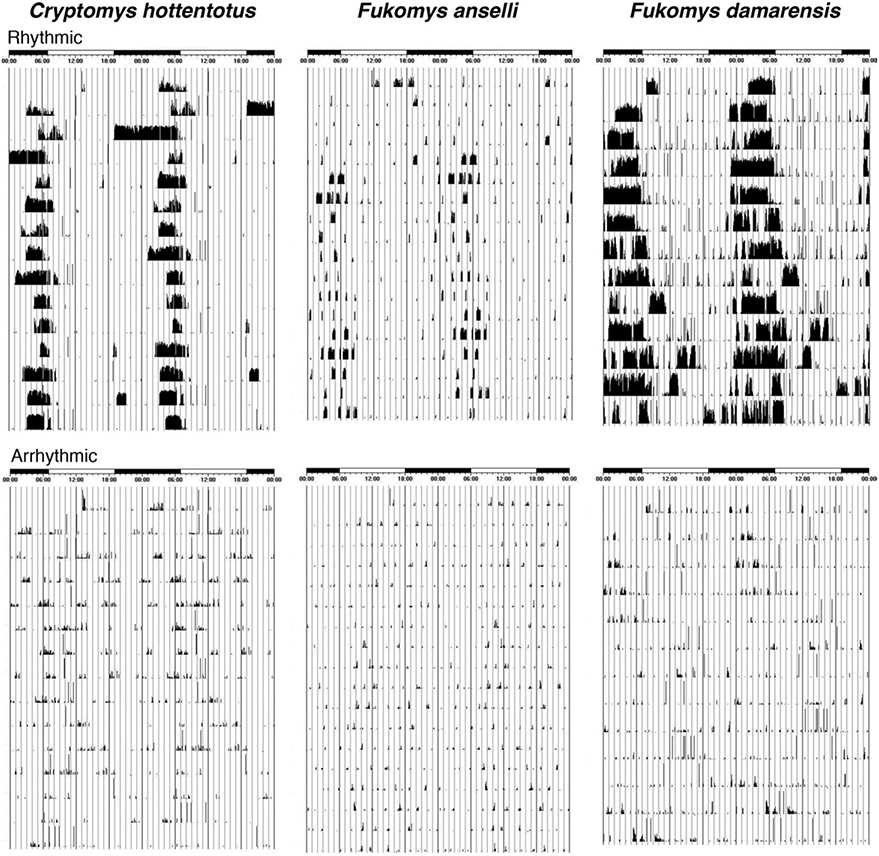
Actigrams illustrating circadian patterns of locomotor activity in rhythmic and arrhythmic individuals from C. hottentotus pretoriae (CHP), F. anselli (FA), and F. damarensis (FD). These actigrams indicate that the rhythmic mole rats have a predictable period of activity during the light period, whereas locomotor activity in the arrhythmic animals is irregular.
Orexin immunohistochemistry
Individuals of each species of mole rat were perfused between 17:00 and 20:00 on three consecutive days, that is, a day per species. The mole rats were euthanazed (Euthanaze, 200 mg sodium pentobarbital/kg, i.p.) and then perfused i.c. The perfusion was initially done with a rinse of 0.9% saline solution at 4 °C, followed by a solution of 4% paraformaldehyde in 0.1 M phosphate buffer (PB) (approximately 1 L/kg of each solution). Brains were removed from the skull and postfixed overnight in 4% paraformaldehyde in 0.1 M PB, and then allowed to equilibrate in 30% sucrose in PB. The brains were blocked and the diencephalon was frozen and sectioned coronally (50-μm section thickness). Tissue from rhythmic and arrhythmic chronotypes of all three species of mole rat in the present study was processed concurrently. A one in two series of stains was made for Nissl and orexin-A immunohistochemistry. Sections used for the Nissl series were mounted on 0.5% gelatine-coated glass slides, cleared in a solution of 1:1 chloroform and absolute alcohol, then stained with 1% Cresyl Violet.
For immunohistochemical staining, the sections were first treated for 30 min with an endogenous peroxidase inhibitor (49.2% methanol: 49.2% of 0.1 PB: 1.6% of 30% H2O2) followed by three 10 min rinses in 0.1 M PB. This was followed by a 2-h preincubation, at room temperature, in a solution (blocking buffer) containing 3% normal goat serum (NGS, Chemicon), 2% bovine serum albumin (BSA, Sigma), and 0.25% Triton X-100 (Merck) in 0.1 M PB. The sections were then incubated in a primary antibody solution containing the appropriately diluted antibody in blocking buffer for 48 h at 4 °C under gentle agitation. To reveal orexinergic neurons, we used the anti-orexin-A antibody AB 3704 (Chemicon, raised in rabbit) at a dilution of 1:1500. This step was followed by three 10-min rinses in 0.1 M PB, after which the sections were incubated in a secondary antibody solution for 2 h. The secondary antibody solution contained a 1:1000 dilution of biotinylated anti-rabbit IgG (BA-1000, Vector laboratories) in 3% NGS and 2% BSA in 0.1 M PB. After three 10-min rinses in 0.1 M PB, the sections were incubated for 1 h in A.B. solution (Vector laboratories) and again rinsed three times. The sections were then treated in a solution of 0.05% diaminobenzidine in 0.1 M PB for 5 min, following which 3 μl of 30% H2O2 was added to the 1 ml of solution in which each section was immersed. Staining development was monitored visually and checked under a low power stereomicroscope. Development was allowed to continue until the immunopositive neurons became readily identifiable from the background stain. Development was arrested by placing the sections in 0.1 M PB, and then rinsed twice more in the same solution. Sections were mounted on glass slides coated with 0.5% gelatine and left to dry overnight. They were then dehydrated in a graded series of alcohols, cleared in xylene, and coverslipped with Depex. Two controls were used in the immunohistochemistry, including the omission of the primary antibody and the omission of the secondary antibody in selected sections, which eliminated staining.
Quantitative analysis
The number of orexinergic (Orx+) positive cells was determined with stereological techniques on a one in two series of sections through the complete hypothalamus in all 18 brains. All analyses were done blind to the rhythmicity status of the individual. A Nikon E600 microscope with three axis motorized stage, video camera, Neurolucida interface, and Stereo-Investigator software (Microbrightfield Corp., Colchester, VT, USA) was used for the stereological counts. In an attempt to achieve the most accurate estimation of Orx+ neurons, a pilot study was first conducted in an individual of each species. The pilot study determined the best counting frame size and grid size, and these parameters were then used for all individuals of each species investigated. A 275×185 μm2 counting frame and a 300×400 μm2 sampling grid were used in each individual. Only orexinergic neurons with clearly visible nuclei were marked in the sampling grids (Table 1). For the calculation of total number of neurons, we measured section thickness in a random sample of 50 sections from each species in the regions where orexinergic neurons were present and used these measurements to calculate the species’ average mounted thickness. The “optical fractionator probe” function of the software computationally determined the total number of Orx+ cell in the hypothalamus of each individual using the following formula:
Where N—was the total estimated neuronal number, Q—was the number of neurons counted, SSF—was the section sampling fraction (in the current study this was 0.5), ASF—was the area sub fraction (this was the ratio of the size of the counting frame to the size of the sampling grid), and TSF—was the thickness sub fraction (this was the ratio of the dissector height relative to 50 μm). To determine TSF, we used the average mounted section thickness calculated for each species (as described earlier in the text, Table 1), subtracted the total vertical guard zones (10 μm) to give dissector height, and used the ratio of dissector height to cut section thickness (50 μm) to provide TSF for each species and applied this to all individuals within a species.
A function in the stereology program called the “nucleator probe” facilitated the estimation of the mean cross-sectional area, volume, and length of the orexinergic positive cells. Only neurons with a distinct nucleus were chosen for analysis. The “nucleator probe” was used in conjunction with the optical fractionator and stereology procedures for systematic random sampling to identify cells (Gunderson, 1988). In total, seven probes were used in the current study, namely: the optical fractionators, optical fractionator using number-weighted section thickness, physical dissector, physical fractionator, Schmitz nearest neighbor, Cavalieri estimator for area and volume, and combination of planes with the optical fractionator for absolute length.
Statistical analysis
In addition, the quantitative values generated using stereological techniques; data for body mass (Mb), brain mass (Mbr) and encephalization quotient (EQ) were compiled for analysis. EQ in each individual was determined using the following formula (from Manger, 2006):
Comparisons were conducted between species (CHP vs. FA, CHP vs. FD and FA vs. FD) and within species (CHP rhythmic vs. CHP arrhythmic, FA rhythmic vs. FA arrhythmic, and FD rhythmic vs. FD arrhythmic). These comparisons were made for Mb, Mbr, EQ, number of Orx+ cell bodies (counts), estimated volume of an Orx+ cell body, estimated cross-sectional area of an Orx+ cell body, and estimated length of an Orx+ cell body. The values obtained for the counts, volume, area, and length of Orx+ cell bodies in each mole rat were divided by the Mb of each individual and the Mbr of each individual thereby creating an index that corrected for Mb and Mbr. These indexes were statistically analyzed between species and within species for the counts, volume, area, and length. The indexes also facilitated comparisons between all rhythmic individuals and all arrhythmic individuals. All data mentioned later in the text are presented as the mean with its standard error. All statistical tests were performed using PAST (Paleontological Statistics, v2.02; Hammer et al., 2001). Initially, the Kruskal–Wallis test was used for the aforementioned comparisons. When significance was observed (P<0.05), multiple Bonferroni-corrected Mann–Whitney U-tests were used as post hoc to the Kruskal–Wallis for the detection of specific differences. P-values for statistically significant differences (again where P<0.05) are provided in the results
RESULTS
In all three species of mole rat, orexinergic immunopositive (Orx+) cell bodies were located within the hypothalamus, as previously reported in all mammals studied to date, including mole rats (Bhagwandin et al., 2011a). Stereological counts of Orx+ cell bodies were found to be statistically significantly different only between the distinct circadian chronotypes of Ansell’s mole rat (FA) and between all rhythmic and arrhythmic individuals of the current study when the counts of Orx+ cell bodies were corrected for Mb. Estimation of cell volume, cross-sectional area, and length showed statistically significant differences between species without correction for Mb and Mbr and statistically significant differences between species when these variables were corrected for Mb and Mbr, although no statistically significant differences were reached for these parameters within species when similar comparisons were made.
Orexinergic cell body distribution
All three species of mole rat expressed Orx+ neurons only within the hypothalamus and were observed as sharing common neuronal locality within the lateral hypothalamic area (LHA), perifornical region (PFR), and the lateral ventral hypothalamic supraoptic area (LVHA), similar to those reported in two other species of mole rat (Bhagwandin et al., 2011a). Thus, there appears to be three distinct clusters of Orx+ neurons in the hypothalamus of the mole rats studied, a large homogeneous cluster spanning the lateral and perifornical regions (the main cluster), a distinct cluster extending into the region of the zona incerta (the zona incerta cluster), and a final cluster in the ventral lateral hypothalamus adjacent to the diminutive optic tracts (the optic tract cluster). The mole rats, from both chronotypes and all three species, exhibited neuronal cell bodies that were morphologically homogenous in all three clusters, that were ovoid in shape, a varying mixture of bi- and multipolar types and that showed no specific dendritic orientation (Figs. 2 and 3).
Fig. 2.
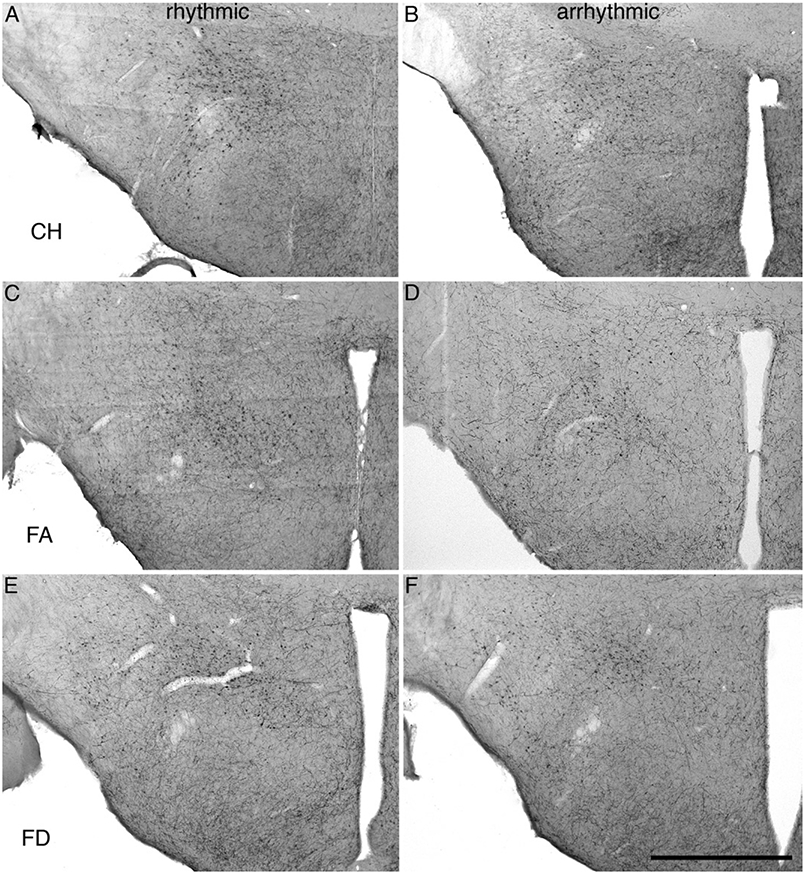
Photomicrographs showing the orexin-A immunoreactive neurons within the hypothalamus of rhythmic and arrhythmic chronotypes of CHP (A, B), FA (C, D), and FD (E, F). Scale = 1 mm and applies to all.
Fig. 3.
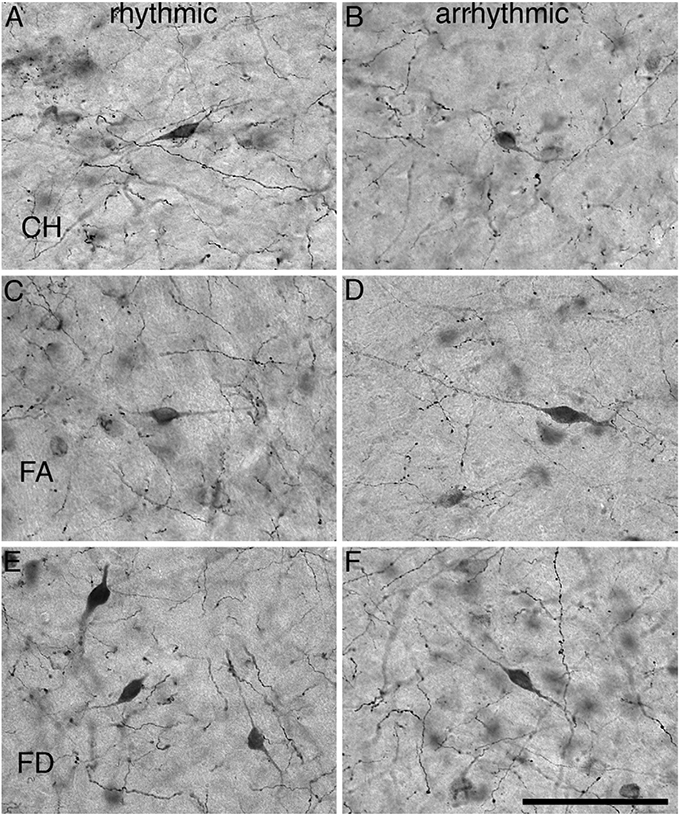
High power photomicrographs showing the morphology of orexin-A immunoreactive neurons within the hypothalamus of rhythmic and arrhythmic chronotypes of CHP (A, B), FA (C, D), and FD (E, F). Scale=100 μm and applies to all.
Comparisons of Mb, Mbr, and EQ
Mean Mb in CHP was 132.8±22.4 g for the rhythmic group and 123±17.6 g for the arrhythmic group. FA had a mean Mb of 90.6±2.8 g for the rhythmic group and 78.5±10.9 g for the arrhythmic group (Table 2). FD had a mean Mb measurement of 121.9±22.7 g for the rhythmic group and 93.7±9.6 g for the arrhythmic group. Mb comparisons between species were only statistically significant (Kruskal–Wallis P-value: 0.0064; Mann–Whitney P-value: 0.008) between CHP and FA where it was observed that CHP had a higher mean Mb (Table 3).
Table 2.
Table presenting data for rhythmic and arrhythmic individuals of C. hottentotus pretoriae, F. anselli, and F. damarensis and the respective weighted means for Mbr, Mb, EQ, estimated cell counts, average estimated volume, average estimated cell area, and average estimated cell length
| Animal ID | Sex | Circadian chronotype |
Body mass (g) |
Brain mass (g) |
EQ | Estimated cell counts |
Average estimated cell volume (μm3) |
Average estimated cell area μm2) |
Average estimated cell length (μm) |
|---|---|---|---|---|---|---|---|---|---|
| C. hottentotus pretoriae | |||||||||
| CHP2 | F | R | 174.5 | 1.6 | 0.6 | 4875 | 1701. 4 | 163.2 | 6.9 |
| CHP8 | F | R | 126.1 | 1.3 | 0.6 | 12,344 | 2438.3 | 198.9 | 7.6 |
| CHP12 | M | R | 97.9 | 1.4 | 0.8 | 9922 | 1936. 5 | 174.4 | 7.2 |
| Mean/weighted mean R | 132.8±22.4 | 1.4±0.09 | 0.6±0.06 | 9047±2200 | 2068.0±98 | 180.9±4.8 | 7.3±0.09 | ||
| CHP1 | M | AR | 110.4 | 1.4 | 0.7 | 11,794 | 2441.3 | 203.9 | 7.8 |
| CHP3 | M | AR | 157.8 | 1.4 | 0.5 | 12,753 | 2520.2 | 204.1 | 7.7 |
| CHP7 | M | AR | 100.7 | 1.7 | 0.9 | 10,095 | 1797. 2 | 167.8 | 7.1 |
| Mean/weighted mean AR | 123.0±17.6 | 1.5±0.10 | 0.7±0.10 | 11,547±777 | 2281.2±116.7 | 193.3±5.9 | 7.5±0.11 | ||
| F. anselli | |||||||||
| FA2 | M | R | 95.9 | 1.2 | 0.7 | 5693 | 2837.0 | 225.3 | 7.9 |
| FA3 | M | R | 86.3 | 1.1 | 0.6 | 5944 | 2875.9 | 223.6 | 8.1 |
| FA7 | M | R | 89.6 | 1.2 | 0.7 | 5719 | 3407.7 | 259.6 | 8.8 |
| Mean/weighted mean R | 90.6±2.8 | 1.2±0.03 | 0.7±0.01 | 5786±80 | 3228.6±172.2 | 247.7±8.1 | 8.6±0.14 | ||
| FA4 | M | AR | 69.3 | 1.2 | 0.8 | 8242 | 3036.8 | 238.6 | 8.4 |
| FA5 | M | AR | 100.1 | 1.1 | 0.6 | 10,686 | 3233.0 | 243.7 | 8.5 |
| FA6 | M | AR | 66.0 | 1.2 | 0.9 | 8969 | 3792.3 | 271.9 | 9.0 |
| Mean/weighted mean AR | 78.5±10.9 | 1.2±0.03 | 0.8±0.09 | 9299±724 | 3337.3±128.7 | 250.7±5.8 | 8.6±0.10 | ||
| F. damarensis | |||||||||
| FD4 | M | R | 76.9 | 1.6 | 1.0 | 8803 | 2428.4 | 204.8 | 7.8 |
| FD6 | M | R | 149.2 | 1.8 | 0.7 | 8024 | 2318.9 | 191.1 | 7.5 |
| FD7 | M | R | 139.7 | 2.0 | 0.8 | 12,723 | 2524.6 | 209.7 | 7.9 |
| Mean/weighted mean R | 121.9±22.7 | 1.8±0.11 | 0.9±0.09 | 9850±1454 | 2429.5±116.2 | 202.4±5.4 | 7.8±0.09 | ||
| FD1 | M | AR | 102.2 | 1.9 | 1.0 | 11,704 | 2289.6 | 198.3 | 7.7 |
| FD8 | F | AR | 104.4 | 1.9 | 1.0 | 9638 | 1988. 0 | 180.3 | 7.4 |
| FD13 | M | AR | 74.5 | 1.7 | 1.0 | 6482 | 2211.6 | 188.8 | 7.4 |
| Mean/weighted mean AR | 93.7±9.6 | 1.8±0.07 | 1.0±0.04 | 9275±1518 | 2163.2±88.9 | 189.3±4.9 | 7.5±0.10 | ||
Table 3.
Table showing results of statistical comparisons between various data sets
| Between species | |||||||
|---|---|---|---|---|---|---|---|
| Body Mass |
Brain Mass |
EQ | Counts | Volume | Area | Length | |
| CHP vs. FA | * CHP | * CHP | – | – | * FA | * FA | * FA |
| CHP vs. FD | – | – | * FD | – | – | – | – |
| FA vs. FD | – | * FD | * FD | – | * FA | * FA | – |
| Within species | |||||||
| CHP R vs. NR | – | – | – | – | – | – | – |
| FA R vs. NR | – | – | – | * AR | – | – | – |
| FD R vs. NR | – | – | – | – | – | – | – |
| Between species corrected for body mass | ||||
|---|---|---|---|---|
| Counts | Volume | Area | Length | |
| CHP vs. FA | – | * FA | * FA | * FA |
| CHP vs. FD | – | – | – | – |
| FA vs. FD | – | * FA | * FA | – |
| Within species corrected for body mass | ||||
| CHP R vs. NR | – | – | – | – |
| FA R vs. NR | * AR | – | – | – |
| FD R vs. NR | – | – | – | – |
| Between species corrected for brain mass | ||||
| Counts | Volume | Area | Length | |
| CHP vs. FA | – | * FA | * FA | * FA |
| CHP vs. FD | – | * CHP | * CHP | * CHP |
| FA vs. FD | – | * FA | * FA | * FA |
| Within species corrected for brain mass | ||||
| CHP R vs. NR | – | – | – | – |
| FA R vs. NR | * AR | – | – | – |
| FD R vs. NR | – | – | – | – |
| All R vs. NR corrected for body mass | |||||
|---|---|---|---|---|---|
| EQ | Counts | Volume | Area | Length | |
| All R vs. NR | – | * AR | – | – | – |
| All R vs. NR corrected for brain mass | |||||
| EQ | Counts | Volume | Area | Length | |
| All R vs. NR | – | – | – | – | – |
Statistical significance as determined using the post hoc Mann–Whitney U and Bonferroni correction factor when P<0.05 (P-values included in-text). Letters in subscript indicate the species or chronotype of mole rat where the higher mean was observed. CHP, C. hottentotus pretoriae; FA, F. anselli; FD, F. damarensis; AR, arrhythmic.
Mean Mbr in CHP measured 1.4±0.09 g for the rhythmic group and 1.5±0.10 g for the arrhythmic group. FA had a mean Mbr measurement of 1.2±0.03 g for the rhythmic group and 1.2±0.03 g for the arrhythmic group. F. damarensis had a mean Mbr measurement of 1.8±0.11 g for the rhythmic group and 1.8±0.07 g for the arrhythmic group (Table 2). Statistically significant differences were noted between the CHP and FA (where the CHP had a higher mean Mbr, Kruskal–Wallis P-value: 0.0039; Mann–Whitney P-value: 0.004) and FA and FD (where FD had a higher mean Mbr, Kruskal–Wallis P-value: 0.0039; Mann–Whitney P-value: 0.004) (Table 3).
Mean EQ in CHP was calculated as 0.6±0.06 for the rhythmic group and 0.7±0.10 for the arrhythmic group. FA had a mean EQ calculation of 0.7±0.01 for the rhythmic group and 0.8±0.09 for the arrhythmic group. FD had a mean EQ calculation of 0.9±0.09 for the rhythmic group and 1 ±0.04 for the arrhythmic group (Table 2). Statistically significant differences were obtained when FD was compared with CHP (where FD had a higher mean EQ, Kruskal–Wallis P-value: 0.02; Mann–Whitney P-value: 0.02) and when FD was compared with FA (where the FD showed a higher mean EQ, Kruskal–Wallis P-value: 0.02; Mann–Whitney P-value: 0.02) (Table 3).
Stereological counts of Orx+ cell bodies
Stereological counts of Orx+ cell bodies in the CHP revealed a mean of 9047±2200 for the rhythmic group and 11,547±777 for the arrhythmic group. Counts from FA revealed a mean of 5786±80 for the rhythmic group and 9299±724 for the arrhythymic group. FD revealed a mean of 9850±1454 for the rhythmic group and 9275±1518 for the arrhythmic group (Fig. 4) (Table 2).
Fig. 4.
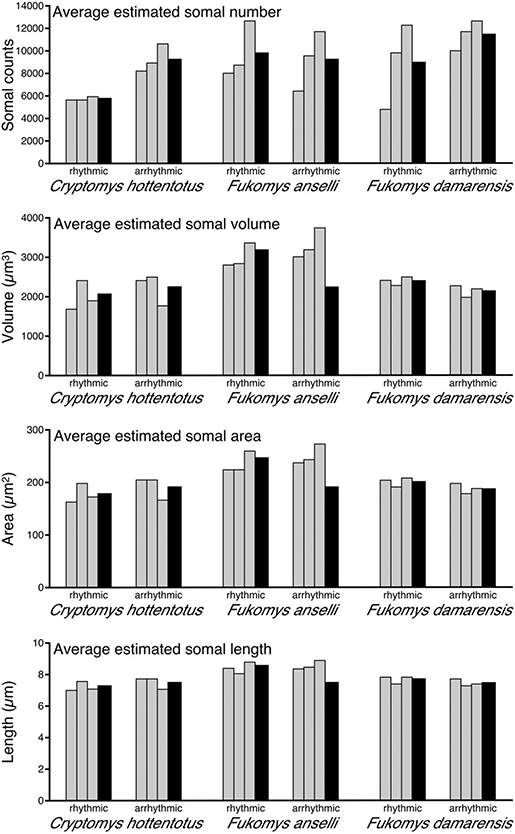
Graphs showing the various parameters of stereological data for arrhythmic and arrhythmic individuals (gray bars) of CHA, FA, and FD and the respective weighted means (black bars). The graphs indicate average estimated values for somal number, volume, area, and length.
Comparisons of Orx+ cell body counts between rhythmic and arrhythmic groups in each species revealed a statistically significant difference only between the circadian chronotypes of FA (where the mean number of Orx+ neurons in the hypothalamus of the arrhythmic group was higher than the rhythmic group, Kruskal–Wallis P-value: 0.008; Mann–Whitney P-value: 0.009). Even though statistical significance was not reached in the other two species investigated, the mean number of Orx+ neurons in the hypothalamus of CHP was higher for the arrhythmic group than the rhythmic group. For FD, stereological counts of Orx+ cell bodies yielded similar totals for both the rhythmic and arrhythmic groups. When the counts of Orx+ cell bodies were corrected for Mb and Mbr, statistically significant differences were noted only between the circadian chronotypes of FA. In both cases, the arrhythmic groups had a higher mean numbers of Orx+ neurons in the hypothalamus than the rhythmic groups (Kruskal–Wallis P-value: 0.0028; Mann–Whitney P-value: 0.003). Additionally, when all rhythmic individuals of all species studied here were compared with all arrhythmic individuals, a statistically significant difference was only reached in counts that had been corrected for Mb (where the arrhythmic group had a higher mean number of Orx+ neurons in the hypothalamus than the rhythmic group, Kruskal–Wallis P-value: 0.02; Mann–Whitney P-value: 0.03) (Table 3).
Stereological estimation of volume, area, and length of Orx+ cell bodies
Stereological estimation of the volume of an Orx+ cell body in CHP presented a weighted mean of 2068±98 μm3 for the rhythmic group and 2281.2±116.7 μm3 for the arrhythmic group. FA presented a weighted mean of 3228.6±172.2 μm3 for the rhythmic group and 3337.3±128.7 μm3 for the arrhythmic group. FD presented a weighted mean of 2429.5±116.2 μm3 for the rhythmic group and 2163.2±88.9 μm3 for the arrhythmic group (Fig. 4) (Table 2). Statistically significant differences were noted: (1) between species (where FA showed a higher mean volume of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.003; Mann–Whitney P-value: 0.0005); (2) when volume was corrected for Mb between species (where FA had a higher mean volume of an Orx+ cell body than CHP, Kruskal–Wallis P-value: 0.004; Mann–Whitney P-value: 0.0008), and where FA had a higher mean volume of an Orx+ cell body than FD (Kruskal–Wallis P-value: 0.004; Mann–Whitney P-value: 0.008); and (3) when volume was corrected for Mbr between species (where FA had a higher mean volume of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.003; Mann–Whitney P-value: 0.005, and where CHP had a higher mean volume of an Orx+ cell body than FD, Kruskal–Wallis P-value: 0.03; Mann–Whitney P-value: 0.03) (Table 3).
Stereological estimation of the cross-sectional area of an Orx+ cell body in the CHP revealed a weighted mean of 180.9±4.8 μm2 for the rhythmic group and 193.3±5.9 μm2 for the arrhythmic group. FA revealed a weighted mean of 247.7±8.1 μm2 for the rhythmic group and 250.7±5.8 μm2 for the arrhythmic group. FD revealed a weighted mean of 202.4±5.4 μm2 for the rhythmic group and 189.3±4.9 μm2 for the arrhythmic group (Fig. 4) (Table 2). Statistically significant differences were noted: (1) between species (where FA showed a higher cross-sectional area of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.003; Mann–Whitney P-value: 0.005); (2) when cross-sectional area was corrected for Mb between species (where FA had a higher cross-sectional area of an Orx+ cell body than CHP, Kruskal–Wallis P-value: 0.004; Mann–Whitney P-value: 0.005, and where FA had a higher cross-sectional area of an Orx+ cell body than FD, Kruskal–Wallis P-value: 0.01; Mann–Whitney P-value: 0.01); and (3) when area was corrected for Mbr between species (where FA had a higher cross-sectional area of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.003; Mann–Whitney P-value: 0.005, and where CHP had a higher cross-sectional area of an Orx+ cell body than FD, Kruskal–Wallis P-value: 0.04; Mann–Whitney P-value: 0.03) (Table 3).
Stereological estimation of the length of an Orx+ cell body in CHP revealed a weighted mean of 7.3±0.09 μm for the rhythmic group and 7.5±0.11 μm for the arrhythmic group. FA revealed a weighted mean of 8.6±0.14 μm for both rhythmic and arrhythmic groups. FD revealed a weighted mean of 7.8±0.09 μm for the rhythmic group and 7.5±0.10 μm for the arrhythmic group (Fig. 4) (Table 2). Statistically significant differences were noted: (1) between species (where FA showed a longer length of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.003; Mann–Whitney P-value: 0.005); (2) when length was corrected for Mb between species (where FA had a longer length of an Orx+ cell body than CHP, Kruskal–Wallis P-value: 0.004; Mann–Whitney P-value: 0.005); and (3) when volume was corrected for Mbr between species (where FA had longer length of an Orx+ cell body than the other two species, Kruskal–Wallis P-value: 0.001; Mann–Whitney P-value: 0.005, and where CHP had a longer length of an Orx+ cell body FD, Kruskal–Wallis P-value: 0.02; Mann–Whitney P-value: 0.03) (Table 3).
DISCUSSION
The initial aim of the current study was to determine whether rhythmic and arrhythmic chronotypes had different numbers of hypothalamic orexin immunopositive neurons with the hypothesis of elevated hypothalamic orexinergic neurons in the arrhythmic chronotypes as a result of the involvement of orexins in the sleep–wake cycle. The results of the current study indicated statistically significant differences between the circadian chronotypes of FA, where the arrhythmic group had higher mean numbers of hypothalamic orexin neurons compared with the rhythmic group. No statistically significant differences in orexinergic neuronal numbers were observed in the other two species studied. A statistically significant difference was noted between all rhythmic and arrhythmic individuals of the current study when the counts of orexin neurons were corrected for Mb.
Orexins, sleep–wake cycle, and circadian rhythmicity
The results of the present study indicated statistically significant differences between circadian chronotypes of FA for the counts of hypothalamic Orx+ neurons when the raw data were compared. In this case, the arrhythmic chronotype consistently displayed higher mean counts of hypothalamic Orx+ neurons than the rhythmic chronotype. A similar tendency, although not statistically significant, was observed when the raw data were compared between the chronotypes of CHP; however, a similar comparison between chronotypes of FD showed that the mean count of Orx+ neurons in the rhythmic group was higher than the rhythmic group, but statistical significance was not reached. The analysis of FA supports the hypothesis of elevated Orx+ neurons in the hypothalamus. Given the previous suggestions for the role of orexins in the sleep–wake cycle (Siegel, 2004; Baumann and Bassetti, 2005), the results of the current study indicate that the arrhythmic chronotypes of FA and possibly CHP would theoretically have an enhanced maintenance of waking (despite the reduced motor activity of the arrhythmic chronotype compared with the rhythmic, as indicated by their actograms, with a net result that possibly enhances vigilance during quiet waking) and a reduced amount of time spent in NREM relative to the rhythmic chronotypes. The study of sleep in rhythmic and arrhythmic chronotypes of the Giant Zambian mole rat (Fukomys mechowii) (Bhagwandin et al., 2011b) has shown that the arrhythmic group statistically spent more time awake and spent less time in NREM compared with the rhythmic group.
It is known that the bifurcated ascending arousal system incorporates first, a pathway to the thalamus which excites thalamic relay neurons that transmit information to the cerebral cortex, and second, a branch that bypasses the thalamus to activate neurons in the LHA, basal forebrain, and cerebral cortex (Saper, 1985; Hallanger et al., 1987; Saper et al., 2001, 2005; Jones, 2003). Orexinergic neurons, with cell bodies restricted to the hypothalamus, have ascending projections to the cerebral cortex and descending projections to the cholinergic (basal forebrain, lateral dorsal tegmental nucleus, pedunculopontine nucleus) and monoaminergic (locus coeruleus, ventral tegmentum area, raphe) nuclear groups of arousal systems (Peyron et al., 1998; Chemelli et al., 1999; Baumann and Bassetti, 2005; Bhagwandin et al., 2011a). The exact role of orexins in REM is unclear, as some studies suggest increased orexinergic activity during REM, whereas others report the contrary (Lee et al., 2005; Mileykovskiy et al., 2005); however, a recent study has postulated that orexin neurons mediate the circadian timing of REM by suppressing REM during the active period (Kantor et al., 2009). In light of the results of the current study, it would appear that the increased numbers of hypothalamic orexinergic neurons observed within FA arrhythmic chronotypes, and potentially CHP, through possible increased activity and the resulting effect on wakefulness and motor activities, may facilitate a higher level of vigilance allowing for extended foraging and feeding activity.
Baumann and Bassetti (2005) proposed that orexins are under control of the SCN and that orexins do not significantly affect circadian rhythmicity. Saper et al. (2005) described a three stage circuitry integrator for circadian rhythms that involve a pathway from the SCN to the sub-paraventricular zone (SPZ) and finally to dorsomedial nucleus of the hypothalamus (DMH). The DMH communicates directly with the LHA, the region with the highest population of orexinergic neurons, and the ventrolateral preoptic nucleus (VLPO), the area primarily concerned with the promotion of slow wave sleep (SWS). The rationale behind the three-stage pathway lies in the observation that in both diurnal and nocturnal animals, the SCN is primarily active during the light period and the VLPO during the dark period (Sherin et al., 1996; Gaus et al., 2002). Therefore, in animals that are nocturnal vs. diurnal, there should be an intervening circuitry that allows the circadian cycle to be set at opposite phases (Saper et al., 2005). It is well known that mole rats have unusual patterns of circadian rhythmicity (Oosthuizen et al., 2003; Lovegrove and Muir, 1996; Lovegrove and Papenfus, 1995) and that mole rats have different neuropeptide populations in the SCN compared with other rodents (Negroni et al., 2003). Therefore, the increased counts of Orx+ neurons in the hypothalamus within FA arrhythmic chronotypes, and potentially CHP, may alter the circadian control of wakefulness in these individuals.
Orexins and appetite regulation
The results of the present study indicated statistically significant differences between circadian chronotypes of FA when the raw data for counts of Orx+ neurons were corrected for Mb and Mbr, where the arrhythmic groups showed higher mean counts than the rhythmic groups. This can be interpreted as the arrhythmic chronotype having a higher number of Orx+ neurons per gram of Mb or Mbr compared with the rhythmic chronotype. Since the initial demonstration of orexin induced hyperphagia in rats (Sakurai et al., 1998), orexins have become synonymous with mechanisms of appetite regulation and increased food intake that is driven by orexin involvement in motivated behavior (Rodgers et al., 2002). Cai et al. (2001) have reported activation of the lateral hypothalamic orexin neurons in response to low blood glucose levels and an empty stomach, signals that indicate replenishment, and concluded that this context-dependency explains the effects associated with orexin activation—increased food intake with delayed onset of satiety and increased wakefulness with vigilance and behavioral activity. It is also known that orexins are involved in appetite regulation through activation of the leptin sensitive neurons of the arcuate nucleus and its subsequent feeding associated signaling to orexinergic neurons of the LH and henceforth interaction between LH and the hypothalamic VMH (Stellar, 1954; Rodgers et al., 2002; Sakurai, 2003, 2005). Therefore, it would seem fair to infer that the elevated counts of hypothalamic orexinergic neurons in the arrhythmic chronotype of FA may yield a greater drive for motivated behavior and theoretically a higher food intake compared with the rhythmic chronotype.
Interestingly though, the results of the present study showed a statistically significant difference for the counts of Orx+ neurons that were corrected for Mb when all rhythmic individuals were compared with all arrhythmic individuals from all three species of mole rats examined. This indicated that the arrhythmic individuals have a higher number of Orx+ neurons per gram of Mb. Given the effect of orexins on motivated behavior and food intake, this may be indicative of a higher Mb in the arrhythmic individuals compared with the arrhythmic individuals. This was not the case, as it was observed in the present study that the mean Mb of the arrhythmic chronotype in each species investigated was lower than that of the rhythmic chronotype. A possible explanation for this could lie in the assessment of the impact of chronic administration of orexin-A on food intake on rats (Haynes et al., 1999; Yamanaka et al., 1999). Both these studies indicated an increased food intake during the light period and a compensatory reduction of food intake during the dark period, and both agree that chronic administration of orexin-A does not alter 24 h food intake or Mb gain. In addition, Lubkin and Stricker-Krongrad (1998) postulated that orexins may increase overall metabolic rate to support increased motor activity; however, Nicolaidis (2006) suggests that overall metabolic rate be divided into resting metabolism and locomotion-related metabolism and based on resting metabolism, a hypometabolic but physically active animal has a higher (total) metabolism than a hypermetabolic animal that remains quiet. This concept supports the observations made in the mole rats assessed in the current study, as it has been previously reported that subterranean mole rats have lower resting metabolic rates than surface dwelling mammals (Bennett and Faulkes, 2000; Zelová et al., 2007, 2009). Furthermore, it has been suggested that sleep duration tracks the metabolism of nutrients in a meal and the size of a meal, with the effect that a higher nutritional content and larger meal size produces a longer sleep duration (Danguir and Nicolaidis, 1980a,b). Despite this, brain metabolism is conserved and independent from that of the periphery, the VMH, dorsomedial nucleus, and paraventricular nucleus, metabolic strategic areas, are capable of reflecting metabolic changes resulting from bodily depletion and repletion (Nicolaidis, 2006). Therefore, it may be possible that a higher peripheral metabolism may result in a lower Mb despite the elevated Orx+ neurons observed in the arrhythmic chronotype compared with the rhythmic chronotype.
Abbreviations:
- CHP
Cryptomys hottenttotus pretoriae
- FA
Fukomys anselli
- FD
Fukomys damarensis
- LH
lateral hypothalamus
- LHA
lateral hypothalamic area
- Mb
body mass
- Mbr
brain mass
- PB
phosphate buffer
- SCN
suprachiasmatic nucleus
- TSF
thickness sub fraction
- VMH
ventromedial nucleus
REFERENCES
- Baumann CR, Bassetti CL (2005) Hypocretins (orexins): clinical impact of the discovery of a neurotransmitter. Sleep Med Rev 9:253–268. [DOI] [PubMed] [Google Scholar]
- Bennett NC, Faulkes CG (2000) African mole-rats: ecology and eusociality. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Bhagwandin A, Fuxe K, Bennett NC, Manger PR (2011a) Distribution of orexinergic neurons and their terminal networks in the brains of two species of African mole rats. J Chem Neuroanat 41:32–42. [DOI] [PubMed] [Google Scholar]
- Bhagwandin A, Gravett N, Oosthuisen MK, Bennett NC, Lyamin OI, Siegel JM, Manger PR (2011b) Sleep and wake in rhythmic and arrhythmic chronotypes of a microphthalmic species of African mole rat (Cryptomys mechowi). Brain Behav Evol 78:162–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XJ, Evans ML, Lister CA, Leslie RA, Arch JR, Wilson S, Williams G (2001) Hypoglycaemia activates orexin neurons and selectively increases hypothalamic orexin-B levels: responses inhibited by feeding and possibly mediated by the nucleus of the solitary tract. Diabetes 50:105–112. [DOI] [PubMed] [Google Scholar]
- Cernuda-Cernuda R, García-Fernández JM, Gordijin MCM, Bovee-Geurts PHM, DeGrip WJ (2003) The eye of the African mole-rat Cryptomys anselli: to see or not to see? Eur J Neurosci 17:709–720. [DOI] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, Fitch TE, Nakazato M, Hammer RE, Saper CB, Yanagisawa M (1999) Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98:437–451. [DOI] [PubMed] [Google Scholar]
- Danguir J, Nicolaidis S (1980a) Cortical activity and sleep in the rat lateral hypothalamic syndrome. Brain Res 185:305–321. [DOI] [PubMed] [Google Scholar]
- Danguir J, Nicolaidis S (1980b) Intravenous infusion of nutrients and sleep in the rat: an ischymetric sleep regulation hypothesis. Am J Physiol 238:E307–E312. [DOI] [PubMed] [Google Scholar]
- Edwards CM, Abusnana S, Sunter D, Murphy KG, Ghatei MA, Bloom SR (1999) The effect of the orexins on food intake: comparison with neuropeptide Y, melanin concentrating hormone and galanin. J Endocrinol 160:R7–R12. [DOI] [PubMed] [Google Scholar]
- Estabrooke IV, McCarthy MT, Ko E, Chou TC, Chemelli RM, Yanagisawa M (2001) Fos expression in orexin neurons varies with behavioral state. J Neurosci 21:1656–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus SE, Strecker RE, Tate BA, Parker RA, Saper CB (2002) Ventrolateral preoptic nucleus contains sleep-active galaninergic neurons in multiple mammalian species. Neuroscience 115:285–294. [DOI] [PubMed] [Google Scholar]
- Gunderson HJ (1988) The nucleator. J Microsc 151:3–21. [DOI] [PubMed] [Google Scholar]
- Gutjahr GH, van Rensburg LJ, Malpaux B, Richter TA, Bennett NC (2004) The endogenous rhythm of plasma melatonin and its regulation by light in the highveld mole-rat (Cryptomys hottentotus pretoriae): a microphthalmic, seasonally breeding rodent. J Pineal Res 37:185–192. [DOI] [PubMed] [Google Scholar]
- Hallanger AH, Levey AI, Lee HJ, Rye DB, Wainer BH (1987) The origins of cholinergic and other subcortical afferents to the thalamus in the rat. J Comp Neurol 262:104–124. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron 4:9pp. [Google Scholar]
- Haynes AC, Jackson B, Overend P, Buckingham RE, Wilson S, Tadayyon M, Arch JR (1999) Effects of single and chronic intracerebroventricular administration of the orexins on feeding in the rat. Peptides 20:1099. [DOI] [PubMed] [Google Scholar]
- Jones BE (2003) Arousal systems. Front Biosci 8:S438–S451. [DOI] [PubMed] [Google Scholar]
- Kantor S, Mochizuki T, Janisiewicz AM, Clark E, Nishino S, Scammell TE (2009) Orexin neurons are necessary for the circadian control of REM sleep. Sleep 32:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Hassani OK, Jones BE (2005) Discharge of identified orexin/ hypocretin neurons across the sleep-walking cycle. J Neurosci 25:6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Faraco J, Kadotani H, Rogers W, Lin X, Qui X, de Jong P, Nishino S, Mignot E (1999) The REM sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98:365–376. [DOI] [PubMed] [Google Scholar]
- Lovegrove BG, Muir A (1996) Circadian body temperature rhythms of the solitary cape mole rat Georychus capensis (Bathyergidae). Physiol Behav 60:991–998. [DOI] [PubMed] [Google Scholar]
- Lovegrove BG, Papenfus ME (1995) Circadian activity rhythms in the solitary cape mole rat (Georychus capensis: Bathyergidae) with some evidence of splitting. Physiol Behav 58:679–685. [DOI] [PubMed] [Google Scholar]
- Lubkin M, Stricker-Krongrad A (1998) Independent feeding and metabolic actions of orexins in mice. Biochem Biophys Res Commun 253:241–245. [DOI] [PubMed] [Google Scholar]
- Manger PR (2006) An examination of cetacean brain structure with a novel hypothesis correlating thermogenesis to the evolution of a big brain. Biol Rev Camb Philos Soc 81:293–338. [DOI] [PubMed] [Google Scholar]
- Mileykovskiy BY, Kiyashchenko LI, Siegel JM (2005) Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron 46:696–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni J, Bennett NC, Cooper HM (2003) Organization of the circadian system in the subterranean mole rat, Cryptomys hottentotus (Bathyergidae). Brain Res 967:48–62. [DOI] [PubMed] [Google Scholar]
- Nemec P, Burda H, Peichl L (2004) Subcortical visual systems of the African mole-rat Cryptomys anselli: to see or not to see? Eur J Neurosci 20:757–768. [DOI] [PubMed] [Google Scholar]
- Nemec P, Cveková P, Benada O, Wielkopolska E, Olkowicz S, Turlejski K, Burda H, Bennett NC, Peichl L (2008) The visual system in subterranean African molerats (Rodentia, Bathyergidae): retina, subcortical visual nuclei and primary visual cortex. Brain Res Bull 75:356–364. [DOI] [PubMed] [Google Scholar]
- Nicolaidis S (2006) Metabolic mechanism of wakefulness (and hunger) and sleep (and satiety): role of adenosine triphosphate and hypocretin and other peptides. Metabolism 55:S24–S29. [DOI] [PubMed] [Google Scholar]
- Oelschläger HH, Nakamura M, Herzog M, Burda H (2000) Visual system labeled by c-Fos immunohistochemistry after light exposure in the “blind” subterranean Zambian mole-rat (Cryptomys anselli). Brain Behav Evol 55:209–220. [DOI] [PubMed] [Google Scholar]
- Oosthuizen MK, Cooper HM, Bennett NC (2003) Circadian rhythms of locomotor activity in solitary and social species of African mole rats (Family: Bathyergidae). J Biol Rhythms 18:481–490. [DOI] [PubMed] [Google Scholar]
- Peyron C, Faraco J, Rogers W, Ripley B, Overeem S, Charnay Y, Nevsimalova S, Aldrich M, Reynolds D, Albin R, Li R, Hungs M, Pedrazzoli M, Padigaru M, Kucherlapati M, Fan J, Maki R, Lammers GJ, Bouras C, Kucherlapati R, Nishino S, Mignot E (2000) A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med 6:991–997. [DOI] [PubMed] [Google Scholar]
- Peyron C, Tighe DK, van den Pol AN, De Lecea L, Heller HC, Sutcliffe JG, Kilduff TS (1998) Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci 18:9996–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers RJ, Ishii Y, Halford JC, Blundell JE (2002) Orexins and appetite regulation. Neuropeptides 36:303–325. [DOI] [PubMed] [Google Scholar]
- Sakurai T (2003) Orexin: a link between energy homeostasis and adaptive behavior. Curr Opin Clin Nutr 6: 353–360. [DOI] [PubMed] [Google Scholar]
- Sakurai T (2005) Roles of orexin/hypocretin in regulation of sleep/wakefulness and energy homeostasis. Sleep Med Rev 9:231–241. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch JR, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M (1998) Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92:573–585. [DOI] [PubMed] [Google Scholar]
- Saper CB (1985) Organization of cerebral cortical afferent systems in the rat. II. Hypothalamocortical projections. J Comp Neurol 237:21–46. [DOI] [PubMed] [Google Scholar]
- Saper CB, Chou TC, Scammell TE (2001) The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci 24:726–731. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J (2005) Hypothalamic regulation of sleep and circadian rhythms. Nature 437:1257–1263. [DOI] [PubMed] [Google Scholar]
- Sherin JE, Shiromani PJ, McCarley RW, Saper CB (1996) Activation of ventrolateral preoptic neurons during sleep. Science 271:216–219. [DOI] [PubMed] [Google Scholar]
- Siegel JM (1999) Narcolepsy: a key role for hypocretins (orexins). Cell 98:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JM (2004) Hypocretin (orexin): role in normal behavior and neuropathology. Annu Rev Psychol 55:125–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellar E (1954) The physiology of motivation. Psychol Rec 61:5–22. [DOI] [PubMed] [Google Scholar]
- Taheri S, Lin L, Austin D, Young T, Mignot E (2004) Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med 1:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal TC, Lai YY, Siegel JM (2007) Hypocretin (orexin) cell loss in Parkinson’s disease. Brain 130:1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, Cornford M, Siegel JM (2000) Reduced number of hypocretin neurons in human narcolepsy. Neuron 27:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal TC, Nienhuis R, Siegel JM (2009) Localized loss of hypocretin (orexin) cells in narcolepsy without cataplexy. Sleep 32:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanItallie TB (2006) Sleep and energy balance: interactive homeostatic systems. Metabolism 55:S30–S35. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Sakurai T, Katsumoto T, Yanagisawa M, Goto K (1999) Chronic intracerebroventricular administration of orexin-A to rats increases food intake in daytime, but has no effect on body weight. Brain Res 849:248–252. [DOI] [PubMed] [Google Scholar]
- Zelová J, Sumbera R, Okrouhlik J, Burda H (2009) Cost of digging is determined by intrinsic factors rather than by substrate quality in two subterranean rodent species. Physiol Behav 99:54–58. [DOI] [PubMed] [Google Scholar]
- Zelová J, Sumbera R, Sedlácek F, Burda H (2007) Energetics in a solitary subterranean rodent, the silvery mole-rat, Heliophobius argenteocinereus, and allometry of RMR in African molerats (Bathyergidae). Comp Biochem Physiol A Mol Integr Physiol 147:412–419. [DOI] [PubMed] [Google Scholar]


