Figure 3.
Construction and validation of the CAROM-ML model
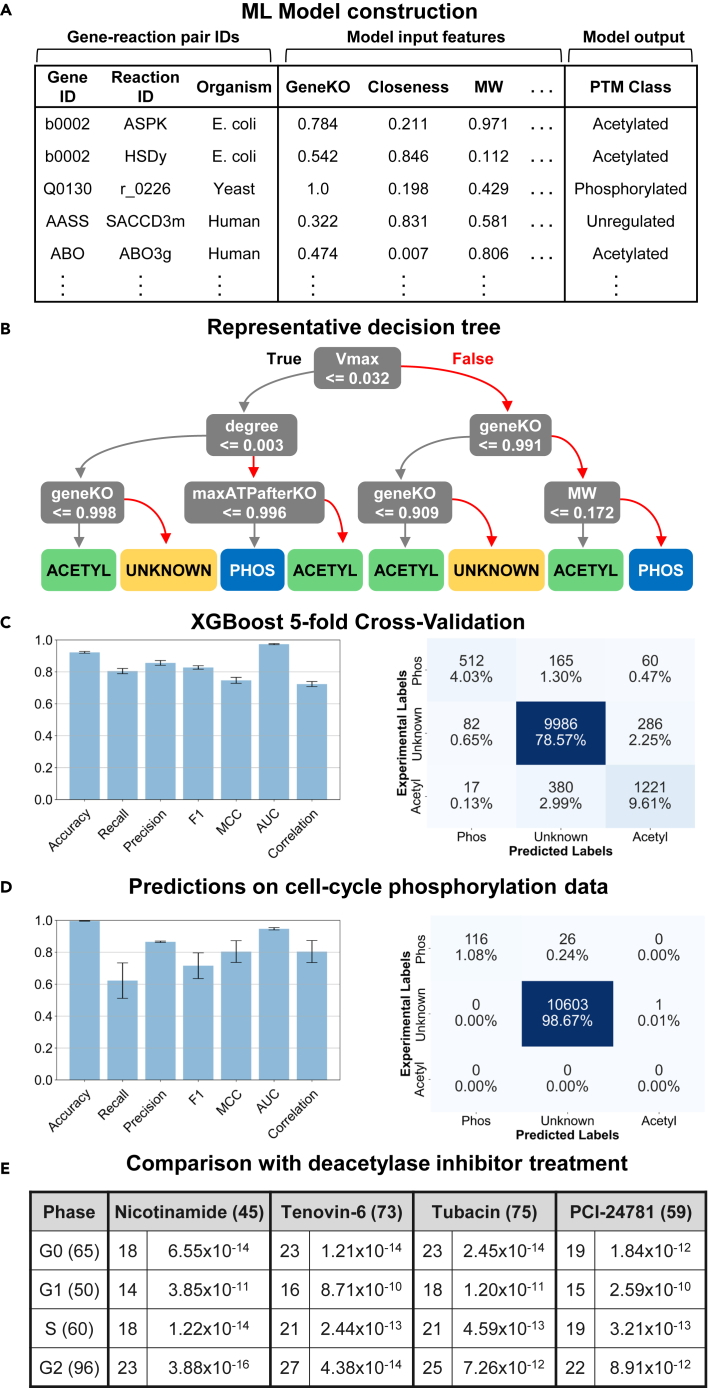
(A) Table of inputs for CAROM-ML. The input features comprise 13 gene, reaction, and enzyme properties. The target column includes the posttranslational modification class. Each gene-reaction pair is marked as either phosphorylated, acetylated, or unknown.
(B) A single decision tree model was built by training on the observations from all organisms, while only using the top 50% most important features as identified in the SHAP analysis. The complexity of the tree was constrained by limiting the tree depth to enable ease of interpretation and visualization. The XGBoost model is made of an ensemble of such decision trees.
(C) The results from the CAROM-ML model from 5-fold cross validation are shown in the bar graph (left) with the 95% confidence intervals represented by the error bars. The cross-validation results are also shown in the confusion matrix.
(D) Comparison of model predictions for the G1, S, and G2 phases of the cell cycle with experimental phospho-proteomics data for those phases. Confusion matrix shows predictions from the main CAROM-ML model, whereas the bar graph shows the standard deviation for five models trained with different random seeds.
(E) Comparison of cell cycle acetylation predictions with experimental acetylomics data from HeLa cells treated with pan-deacetylase inhibitors. The number of unique acetylated genes for each group are displayed in parentheses. Within the table, the number of overlapping genes between each phase and drug is shown, along with the p value of the hypergeometric test.