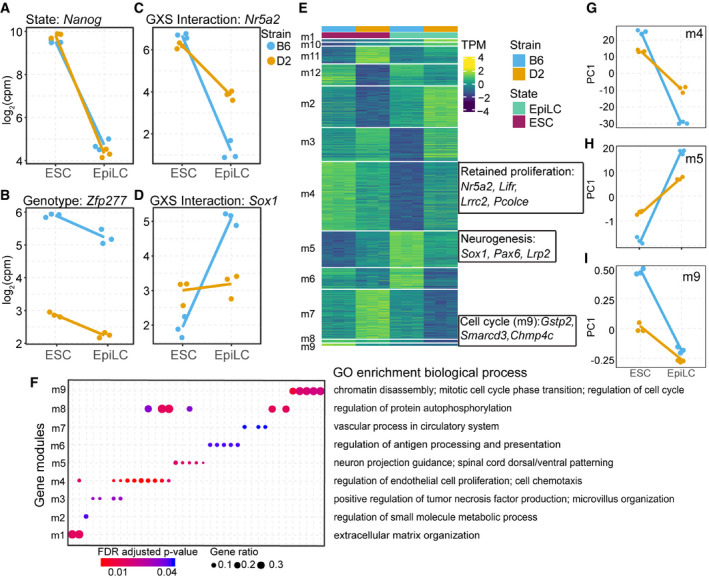
-
A–D
Example expression patterns for select genes identified by applying a general linear model (glm) including state, genotype, and interaction terms. Dots represent individual biological replicates (N = 3); lines highlight changes in mean values for replicates between each state (log2FC > 1 and FDR < 0.05). (A) State‐dependent expression is exemplified by Nanog, which showed no difference between strains. (B) Expression of Zfp277 exemplifies strain dependence being consistently higher in B6. (C, D) A significant genotype x state (GXS) interaction was identified for Nr5a2 (C, a marker for pluripotency) and Sox1 (D, a marker for neuronal differentiation).
-
E
Heatmap of transcript abundances for individual genes (rows) representing 12 expression modules detected by EBseqHMM filtered for genes with a significant GXS glm interaction. Genes with observed functional similarity within modules are highlighted. Nr5a2, Lifr, and other pluripotency genes shared the same expression path (m4) that decreases more significantly in B6, retained expression in D2, when ESCs are differentiated to EpiLCs. Sox1, Pax6, and other neurogenesis genes shared an expression path (m5) that is significantly upregulated in B6 EpiLCs. Gstp2, Smarcd3, and other cell cycle regulation genes shared an expression path (m9) that is significantly upregulated in B6 ESCs.
-
F
Gene ontology (GO) enrichment of biological processes (FDR‐adjusted P‐value < 0.05) identified for 9 out of 12 expression modules (m1‐9). Each column represents a GO term. Highlighted terms are indicated to the right. GO terms associated with proliferation (m4), neurogenesis (m5), and cell cycle regulation (m9) indicate genetic control of exit from pluripotency and early lineage priming.
-
G–I
Summarized differential module behavior between B6 and D2 (3 biological replicates) upon transition from ESCs to EpiLCs represented by PC1 of transcript abundance for all genes within each module. (G) D2 retained proliferation in EpiLCs. (H) Upregulation of neurogenesis in B6 EpiLCs. (I) Increased regulation of cell cycle M phase in B6 ESCs.