Figure 6. Validation of Chr 4 QTL on gene expression and chromatin accessibility implicate KRAB‐ZFPs in trans‐regulation.
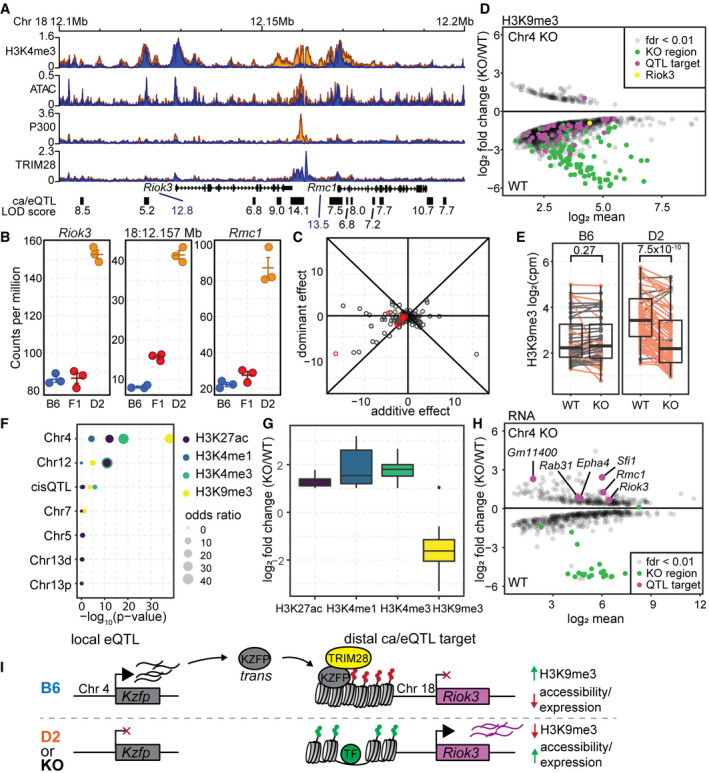
- Coverage profile for H3K4me3, ATAC, P300, and TRIM28 ChIP at a target locus for the QTL on Chr 4 for both chromatin accessibility (black boxes) and gene expression (Riok3 and Rmc1). LOD scores are listed under each target locus.
- Gene expression (Riok3 and Rmc1) or chromatin accessibility (18:12.157 Mb) level measured in B6, D2, and (BxD)F1 hybrid mESCs. For this locus, both gene expression and chromatin accessibility are dominantly repressed in the F1 (mean ± SEM).
- Scatterplot of the additive versus dominant effect for each Chr 4 caQTL target locus. Points along the diagonal lines indicate dominant effects (red—caQTL targets found at the Riok3/Rmc1 locus.
- MA plot of significant (FDR < 0.01) differential H3K9me3 level between wild‐type (B6) mESCs and those with a 2.47‐Mb deletion encompassed by the Chr 4 QTL (n = 2 replicates). Regions that overlap Chr 4 caQTL targets are indicated in magenta (Chr18:12.157 Mb locus from (A) in yellow).
- Box‐and‐whisker plot indicating change in H3K9me3 levels between WT and Chr 4 KO mESCs grouped by whether chromatin accessibility is higher when the QTL is B6 (left) or D2 (right). Dots represent mean values from biological duplicate experiments, and lines connect means. Orange indicates loci for which H3K9me3 signal decreases in the KO (P‐value indicated on top, two‐sided paired Wilcoxon’s test).
- LOLA analysis of significant enrichment between sites that change between WT and Chr 4 KO mESCs and overlap QTL hotspot targets. For H3K4me3, H3K4me1, and H3K27ac, there was only one ChIP experiment performed, and therefore, all sites with a log2 fold change ≥ 1 were used for overlap enrichment.
- Box‐and‐whisker plot indicating direction of change for histone modifications between WT and Chr 4 KO cells.
- MA plot similar to (D) showing change in gene expression between WT and Chr 4 KO mESCs (n = 2). Chr 4 QTL targets are indicated in magenta and increase expression in the KO.
- Model of trans‐regulation of chromatin state for the Chr 4 QTL. Local variation at the Chr 4 QTL results in differential expression of KZFPs, leading to decreased expression of distal genes through recruitment of TRIM28 and formation of heterochromatin (H3K9me3).
Data information: In the boxplot, the central lines represent medians, the box represents the first and third quartile, and the whiskers extend 1.5 times the interquartile range.
