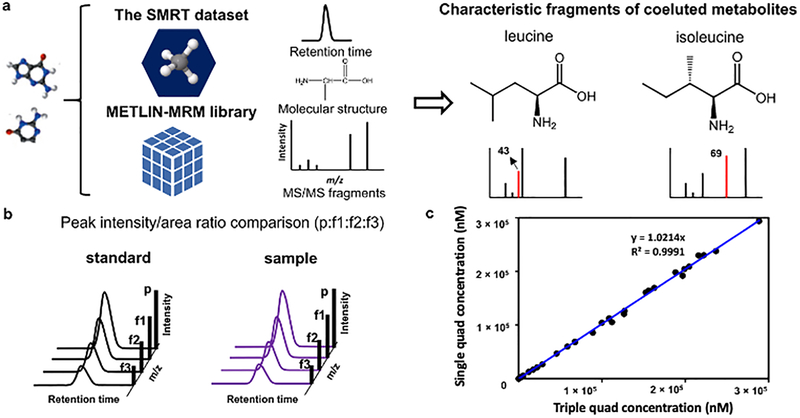
Figure 2.
Single and triple quadrupole characteristic fragments of co-eluting metabolites can be selected using retention time data generated from the SMRT dataset and fragmentation data from the METLIN-MRM library (e.g., (a) leucine and isoleucine). This data can be used to monitor the specificity of quantifier based on (b) peak intensity ratios, for example. Concentrations of 50 metabolites calculated using single quadrupole monitoring aligned well with the results calculated using a triple quadrupole generated from identical cell sample (analytical standards were spiked into the cell sample at varying concentrations if the metabolites were not detected in the cell sample) (c). p: precursor ion; f1, f2 and f3: fragment ion 1, 2, and 3, respectively.